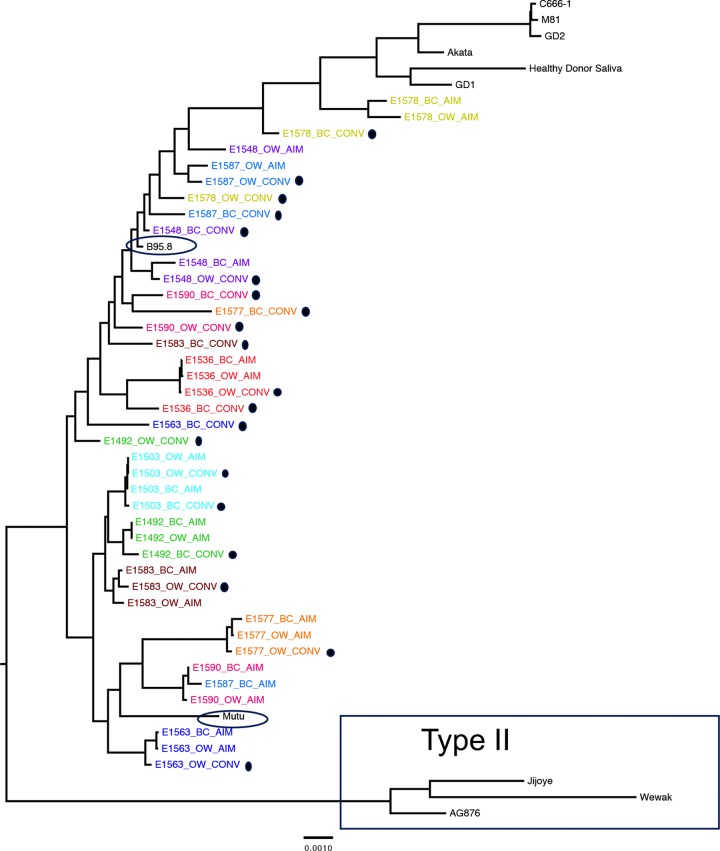
FIG 2.
Forty full-length EBV viral genomes sequenced from 10 patients align almost exclusively with type I reference strains. EBV sequences are labeled by source (peripheral blood B cells, BC; oral wash, OW) and timing of sample (AIM, within 2 weeks of diagnosis of acute infectious mononucleosis; CONV, at least 6 months postinfection [highlighted with black dots]) during primary EBV infection. A maximum-likelihood, midpoint-rooted phylogeny is shown, with each patient represented independently by color. EBV type II sequences are boxed. All other sequences are derived from EBV type I. Sequences from the cohort are most closely related to B95.8 and Mutu isolates (circled) and least related to Asian EBV type I isolates (GD1, Akata, GD2, M81, C666-1, and saliva sample from a healthy Asian donor), shown at the top of the tree.