ABSTRACT
Since its emergence in 2013, the H7N9 low-pathogenic avian influenza virus (LPAIV) has been circulating in domestic poultry in China, causing five waves of human infections. A novel H7N9 highly pathogenic avian influenza virus (HPAIV) variant possessing multiple basic amino acids at the cleavage site of the hemagglutinin (HA) protein was first reported in two cases of human infection in January 2017. More seriously, those novel H7N9 HPAIV variants have been transmitted and caused outbreaks on poultry farms in eight provinces in China. Herein, we demonstrate the presence of three different amino acid motifs at the cleavage sites of these HPAIV variants which were isolated from chickens and humans and likely evolved from the preexisting LPAIVs. Animal experiments showed that these novel H7N9 HPAIV variants are both highly pathogenic in chickens and lethal to mice. Notably, human-origin viruses were more pathogenic in mice than avian viruses, and the mutations in the PB2 gene associated with adaptation to mammals (E627K, A588V, and D701N) were identified by next-generation sequencing (NGS) and Sanger sequencing of the isolates from infected mice. No polymorphisms in the key amino acid substitutions of PB2 and HA in isolates from infected chicken lungs were detected by NGS. In sum, these results highlight the high degree of pathogenicity and the valid transmissibility of this new H7N9 variant in chickens and the quick adaptation of this new H7N9 variant to mammals, so the risk should be evaluated and more attention should be paid to this variant.
IMPORTANCE Due to the recent increased numbers of zoonotic infections in poultry and persistent human infections in China, influenza A(H7N9) virus has remained a public health threat. Most of the influenza A(H7N9) viruses reported previously have been of low pathogenicity. Now, these novel H7N9 HPAIV variants have caused human infections in three provinces and outbreaks on poultry farms in eight provinces in China. We analyzed the molecular features and compared the relative characteristics of one H7N9 LPAIV and two H7N9 HPAIVs isolated from chickens and two human-origin H7N9 HPAIVs in chicken and mouse models. We found that all HPAIVs both are highly pathogenic and have valid transmissibility in chickens. Strikingly, the human-origin viruses were more highly pathogenic than the avian-origin viruses in mice, and dynamic mutations were confirmed by NGS and Sanger sequencing. Our findings offer important insight into the origin, adaptation, pathogenicity, and transmissibility of these viruses to both poultry and mammals.
KEYWORDS: emergence, adaptation, highly pathogenic H7N9 influenza virus, multiple basic amino acids
INTRODUCTION
Avian influenza viruses (AIVs) have posed a significant threat to public health for decades. At least five subtypes of AIV (H5, H6, H7, H9, and H10) have been shown to infect humans in China. Clearly, the continued evolution of AIVs might lead to the emergence of a potentially virulent and highly transmissible AIV variant among human populations.
Human infections with low-pathogenic AIV (LPAIV) H7N9 were first reported in March 2013 in China. Since then, it has continued to exist in China, causing five waves of human infection (1–5). As of 25 July 2017, 1,557 human cases and 605 deaths have been confirmed in 24 provinces, with a steep increase in cases occurring during late 2016 and early 2017 (6). The number of human infections with the H7N9 virus gradually decreased in waves 3 (October 2014 to September 2015) and 4 (October 2015 to September 2016). During this period and despite frequent reassortment (7), no genetic variants associated with human influenza virus-like receptor binding preference, aerosol transmission, or increased pathogenicity were identified. However, during the fifth wave of human infections, from 20 December 2016 to the present, an upsurge in the number of human H7N9 virus infections has been seen in China (6).
Previous studies have shown that LPAIV could become highly pathogenic via the insertion of multiple basic amino acids at the cleavage site of the hemagglutinin (HA) protein, which have been found in H5 and H7 subtype AIVs. Strikingly, some H7N9 variants isolated from humans during January 2017 in Guangzhou City, Guangdong Province, China, possessed an insertion of 4 amino acids, PKRKRTA(R/G), including multiple basic amino acids (underlined), at the cleavage site of the HA protein, regarded as a marker of high pathogenicity of influenza A viruses for chickens (8, 9). More seriously, cases of human H7N9 HPAIV infection have been confirmed in Guangxi and Hunan Provinces in China (6). During our routine surveillance for AIV in live poultry markets (LPMs) in Guangdong Province (2013 to 2017), we also identified as early as mid-2016 H7N9 variants with the insertion of multiple basic amino acids at the cleavage site of HA. When we paid closer attention, it was found that those novel H7N9 HPAIV variants have been transmitted and have caused outbreaks on nine poultry farms in eight provinces (10). To explore the genesis and evolution of the HPAIV H7N9 mutants, we performed a genetic and phylogenetic analysis of all of the HPAIVs along with the LPAIVs isolated from poultry and humans. Animal experiments were then performed to compare the pathogenicity of the avian- and human-origin H7N9 HPAIVs in chickens, ducks, and mice.
RESULTS
Unusual cleavage sites in the HA of the novel H7N9 HPAIV variants.
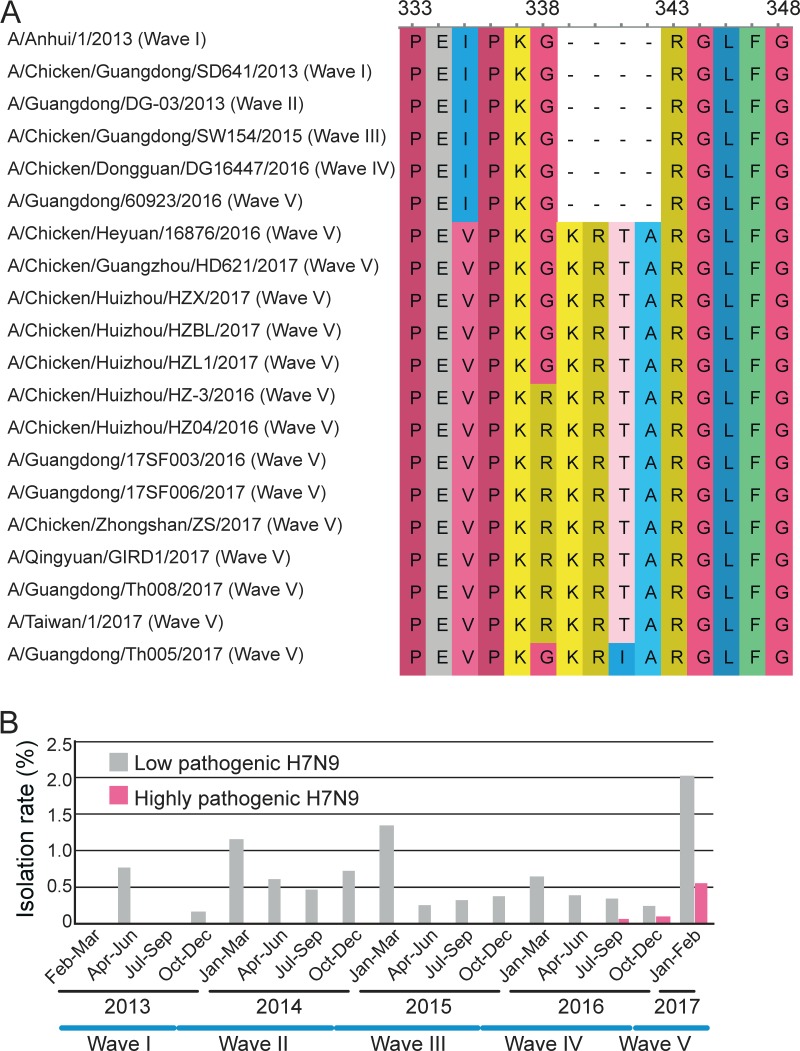
To understand the genesis, prevalence, and pathogenicity of the novel H7N9 HPAIV variants, we developed a real-time reverse transcription-PCR (rRT-PCR) assay (11). We then performed a retrospective analysis of 128 H7N9-positive avian samples collected from 2013 to 2017 by use of this rRT-PCR assay and confirmed the HA cleavage site sequence through Sanger sequencing. Our samples covered all five H7N9 infection waves and 10 counties in Guangdong Province (see Table S1 in the supplemental material). Strikingly, this surveillance identified viruses with three types of unusual HA cleavage sites—the motifs PKGKRTA(R/G), PKRKRTA(R/G), and PKGKRIA(R/G) (Fig. 1A)—from chickens sampled in LPMs and human cases from Guangdong Province. The first two motifs have one nucleotide (G-to-A) difference that leads to an additional amino acid change (R) in the second motif, which also characterized the human isolates recently reported in Guangdong. The third motif contains a C-to-T nucleotide substitution causing an amino acid change from T to I compared to the sequence of the first motif. The earliest highly pathogenic virus (A/Chicken/Heyuan/16876/2016) with the PKGKRTA(R/G) motif was isolated from a swab specimen from a chicken in Heyuan City during wave 4 (24 July 2016). Four additional viruses (three from Huizhou City and one from Guangzhou City) identified in January and February 2017 in wave 5 carried the same insertion (Fig. 1A). The second KRKRTA(R/G) motif was first identified in Huizhou on 28 December 2016 and subsequently in other cities (Fig. 1A). In addition, on the basis of our surveillance data, the rate of isolation of the novel H7N9 HPAIV variant seemingly increased during 2017 (Fig. 1B). Notably, our surveillance also revealed that the HPAIV variants with KRKRTA(R/G) and PKGKRIA(R/G) have also caused human infections (Fig. 1A).
FIG 1.
Representative variants of H7N9 AIVs from Guangdong Province, China. (A) Alignment of the HA cleavage site from the five waves of H7N9 infection. Position numbers are shown according to the sequence of the highly pathogenic H7N9 variant A/Chicken/Heyuan/16876/2016. Wave numbers are marked wave I to wave V. (B) Frequency of isolation of H7N9 viruses from chickens and ducks recovered from 2013 to 2017 in Guangdong Province. The time ranges of the five waves are shown.
Origin of the novel H7N9 HPAIV variants.
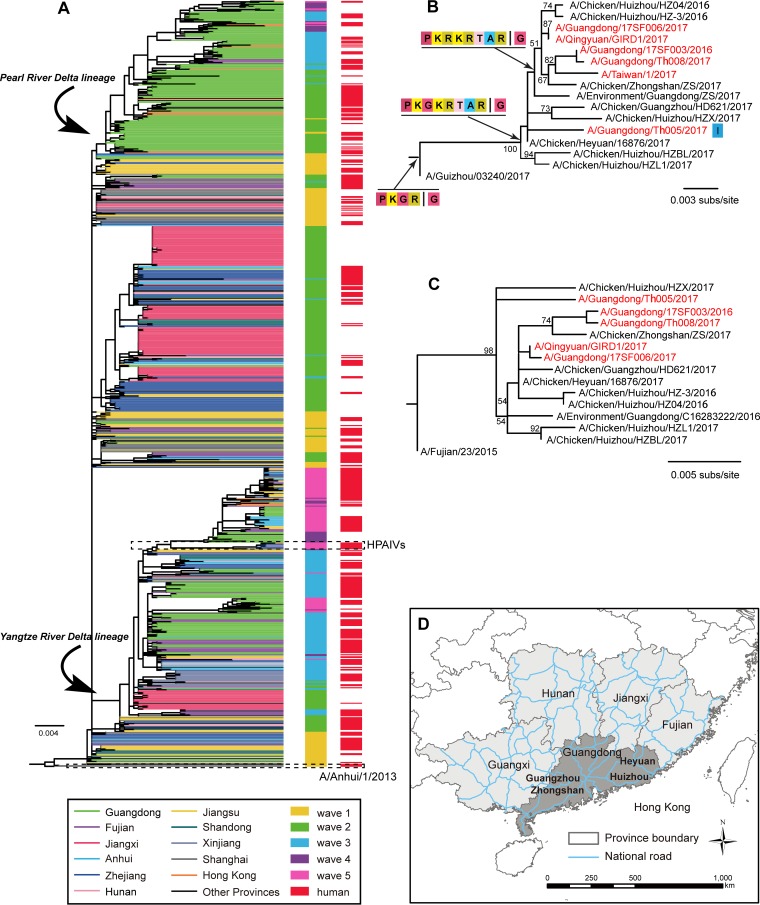
To trace the evolutionary origin of the novel H7N9 HPAIV variants, we performed a phylogenetic analysis of all 96 H7N9 viruses characterized here and those available from GenBank (https://www.ncbi.nlm.nih.gov/GenBank/) and GISAID (platform.gisaid.org). The HA and neuraminidase (NA) phylogenies (Fig. 2, S1, and S2) revealed that the so-called Yangtze River Delta lineage and the Pearl River Delta lineage of H7N9 cocirculated through waves 2 to 5 (4), with the former lineage being dominant in and widely distributed throughout China. H7N9 LPAIV strains of this lineage were repeatedly introduced into or/and reemerged in Guangdong in waves 3, 4, and 5 (Fig. 2A). Importantly, the novel H7N9 HPAIV variants characterized by the insertion of multiple basic amino acids (Fig. 2B) present in both poultry and humans from mainland China grouped together and formed a distinct cluster in both the HA and NA gene trees (Fig. 2B and C), indicative of a single origin. We also found that viruses possessing the motif PKRKRTA(R/G) were farther from the root than strains possessing the motif PKGKRTA(R/G), indicative of additional nucleotide substitutions (Fig. 2B).
FIG 2.
Evolutionary history of H7N9 AIV. (A) Phylogenetic tree of the H7N9 HA gene. Viruses isolated from different provinces and waves are distinguished by colors. Human strains are also highlighted. All branch lengths are scaled according to the number of substitutions (subs) per site. (B) HA gene tree revealing a single cluster of highly pathogenic H7N9 variants isolated from Guangdong Province. Cleavage sites are shown at the nodes. I, a mutation (T to I) in the cleavage site of the strain A/Guangdong/Th005/2017. (C) NA gene tree of the same cluster of highly pathogenic variants for which the results are shown in panel B. (D) A map showing the region of southern China and the major national roads.
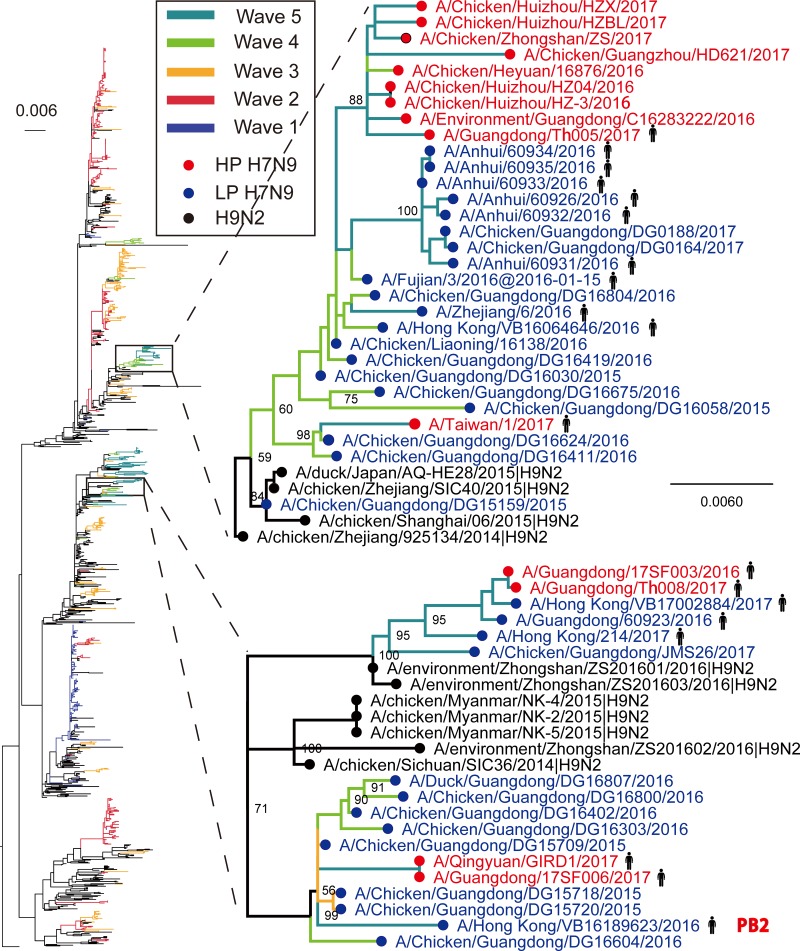
In contrast to the HA and NA genes, the internal genes of the novel H7N9 HPAIV variants in wave 5 characterized here did not cluster together (Fig. S3 to S8). Rather, they were scattered across the tree and formed several clusters (Fig. S3 to S8). For example, in the PB2 phylogeny, the novel H7N9 HPAIV strains formed two independent clusters, both of which were closely related to the H7N9 LPAIV strains from humans and chickens in waves 4 and 5 (Fig. 3). In the trees generated with other internal genes, the novel HPAIV strains also clustered with H7N9/H9N2 LPAIVs circulating locally. These observations suggest that, similar to the findings for H7N9 LPAIVs, the novel H7N9 HPAIV variants have undergone complex reassortment with H7N9/H9N2 LPAIVs and multiple genotypes are cocirculating in poultry. Hence, both H7N9 HPAIV and H7N9 LPAIV recruited internal genes from the same influenza virus gene pool via reassortment.
FIG 3.
Phylogenetic tree of the PB2 gene sequences. The scale bar represents the number of nucleotide substitutions per site. The highly pathogenic H7N9 variants reported in the present study are marked in red, and the remaining H7N9 viruses are marked in blue. The H9N2 AIVs are marked in black. The branches are colored according to the date on which the virus was isolated.
Molecular characterization.
The Q226L mutation in the HA protein is favored for adaptation to mammals and is required for the receptor binding ability. All of the novel HPAIV H7N9 variants from poultry and humans possessed the 226Q mutation, while LPAIVs always possessed 226L. All H7N9 viruses, including low-pathogenic and highly pathogenic viruses isolated from humans and poultry, had the S31N mutation in the M2 protein, which reduced the sensitivity of the virus to amantadine. Importantly, three of four human HPAIV strains carried the R292K mutation in the NA, while no poultry-source H7N9 HPAIVs had this drug resistance mutation. In addition, three of the four human HPAIVs carried the E627K mutation, two viruses had an A588V mutation, and two viruses possessed the K526R mutation all of these mutations are associated with adaptation to mammals. None of the HPAIV variants sampled from chickens carried these mutations for adaptation to mammals (Table S2).
Pathogenesis in animals.
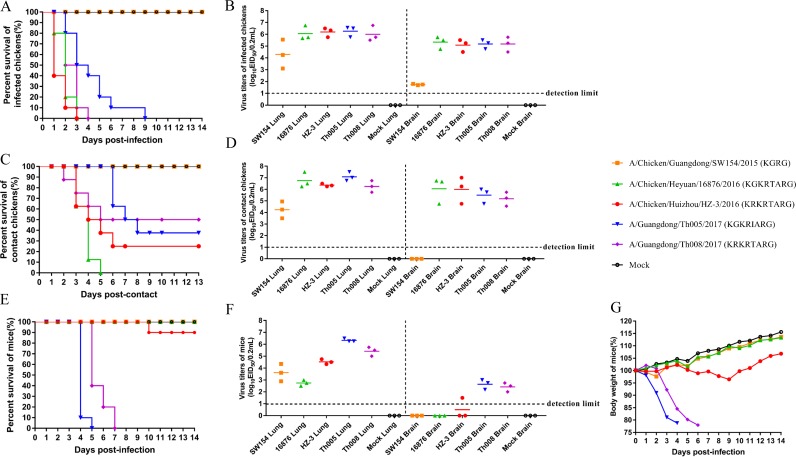
To investigate the pathogenesis of the variants in chickens, A/Chicken/Guangdong/SW154/2015, which does not contain the cleavage site with the insertion of multiple basic amino acids, and the four viruses A/Chicken/Heyuan/16876/2016, A/Chicken/Huizhou/HZ-3/2016, A/Guangdong/Th005/2017, and A/Guangdong/Th008/2017, which contain the cleavage site with the insertion of multiple basic amino acids, were inoculated into 6-week-old specific-pathogen-free (SPF) chickens to perform the intravenous pathogenicity test (IVPI), which resulted in values of 0, 2.92, 3.00, 2.33, and 2.41, respectively (Table 1). Hence, both avian- and human-origin H7N9 variants with the cleavage site with the insertion of multiple basic amino acids were highly pathogenic in chickens.
TABLE 1.
Virus isolation, seroconversion results, and results of IVPI in chicken experiment
| Virus and means of exposure | No. of chickens from which virus was isolated/total no. of chickens tested on: |
No. of chickens surviving on 14 dpi that seroconverted/total no. of chickens | IVPI result | Cleavage site sequence | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 dpi |
5 dpi |
7 dpi |
9 dpi |
||||||||
| Trachea | Cloaca | Trachea | Cloaca | Trachea | Cloaca | Trachea | Cloaca | ||||
| A/Chicken/Guangdong/SW154/2015 | 0 | KGRG | |||||||||
| Infected | 10/10 | 10/10 | 3/7 | 7/7 | 4/7 | 6/7 | 4/7 | 7/7 | |||
| Contact | 8/8 | 8/8 | 7/8 | 8/8 | 4/5 | 5/5 | 3/5 | 2/5 | 5/5 | ||
| A/Chicken/Heyuan/16876/2016 | 2.92 | KGKRTARG | |||||||||
| Infected | 2/2 | 2/2 | |||||||||
| Contact | 2/8 | 1/8 | 5/5 | 5/5 | |||||||
| A/Chicken/Huizhou/HZ-3/2016 | 3.00 | KRKRTARG | |||||||||
| Infected | 1/1 | 1/1 | |||||||||
| Contact | 1/8 | 1/8 | 2/5 | 2/5 | 1/3 | 1/3 | 0/2 | 0/2 | 2/2 | ||
| A/Guangdong/Th005/2017 | 2.33 | KGKRIARG | |||||||||
| Infected | 4/8 | 3/8 | 3/4 | 2/4 | 1/1 | 1/1 | |||||
| Contact | 0/8 | 0/8 | 3/8 | 2/8 | 3/7 | 3/7 | 0/3 | 0/3 | 3/3 | ||
| A/Guangdong/Th008/2017 | 2.41 | KRKRTARG | |||||||||
| Infected | 4/5 | 3/5 | |||||||||
| Contact | 2/8 | 1/8 | 1/6 | 1/6 | 0/4 | 0/4 | 0/4 | 0/4 | 4/4 | ||
| Mock infection | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0 | |
Chickens inoculated intranasally with the four HPAIVs had severe clinical outcomes and gross lesions at necropsy, but chickens challenged with the A/Chicken/Guangdong/SW154/2015 virus did not show any clinical signs during the experimental period, similar to the findings for control chickens. All chickens infected with A/Chicken/Heyuan/16876/2016 and A/Chicken/Huizhou/HZ-3/2016 died within 3 days postinoculation (dpi), and all chickens inoculated with A/Guangdong/Th008/2017 died within 4 dpi (Fig. 4A). Interestingly, although half of the chickens infected by A/Guangdong/Th005/2017 died within 3 dpi, the remainder died in the next 6 days, indicative of pathogenicity lower than that of the other highly pathogenic strains tested here. The organs, including heart, lung, kidney, brain, spleen, and liver, of the first three dead chickens in each group were positive for H7 AIV, and the virus titers in the lungs and brains were high and not significantly different from those of the A/Chicken/Guangdong/SW154/2015 virus in each group (Fig. 4B). We also compared the transmissibility of these viruses among chickens. All of the chickens exposed to A/Chicken/Heyuan/16876/2016 by direct contact died within 5 days; however, 25%, 37.5%, and 50% of the chickens exposed to A/Chicken/Huizhou/HZ-3/2016, A/Guangdong/Th005/2017, and A/Guangdong/Th008/2017 by direct contact survived, respectively, reflecting valid transmissibility in chickens (Fig. 4C). Moreover, the virus titers in the lungs and brains of the dead chickens were high, approximately 106 log10 50% egg infective dose (EID50)/0.2 ml (Fig. 4D). In addition, virus shedding was also detected in tracheal and cloacal swab specimens at 3, 5, 7, 9, 11, and 13 dpi, but none of the surviving chickens exposed to HPAIV showed virus shedding at the different sampling time points and no seroconversion on 14 dpi; however, all of the chickens in the group infected with the A/Chicken/Guangdong/SW154/2015 virus seroconverted (Table 1). The virus sequences in the lung tissue samples from infected chickens and chickens exposed by direct contact were sequenced using next-generation sequencing (NGS) on an Illumina HiSeq 2000 sequencer, and no dynamic mutations were detected. We intranasally infected 4- to 5-week-old female BALB/c mice (weight, 14 to 16 g) with 106 EID50 of each virus. Mice infected with human viruses A/Guangdong/Th005/2017 and A/Guangdong/Th008/2017 showed signs of illness and weight loss starting from 3 dpi and died in 5 or 7 days (Fig. 4E). However, mice infected with avian-origin viruses (i.e., A/Chicken/Guangdong/SW154/2015, A/Chicken/Heyuan/16876/2016, and A/Chicken/Huizhou/HZ-3/2016) did not display obvious clinical signs or weight loss, although mice infected with A/Chicken/Huizhou/HZ-3/2016 showed a slight weight loss (<5%) at about 9 dpi (Fig. 4G), and one mouse (1/10) died at 10 dpi (Fig. 4E). In addition, all four tested viruses could replicate efficiently in the lungs of the infected mice by 4 dpi. There was no difference in the titers of the avian-origin LPAIVs and the avian-origin HPAIVs in the mouse lungs (P > 0.05), but the titers of the human-origin HPAIVs were obviously higher than those of the LPAIVs (0.01 < P ≤ 0.05). The titers of the two human-origin viruses in mouse brains were significantly different from those of the three avian-origin viruses (0.0001 < P ≤ 0.001) (Fig. 4F). In addition, the two human-origin viruses could be detected in the mouse brains, whereas the two avian-origin viruses could not (Fig. 4F). All of the other mice in the groups infected with the three avian-origin viruses recovered and seroconverted at the end of the experiment. The PB2 E627K mutation was found by Sanger sequencing at necropsy at 4 dpi in two mice in the group infected with the A/Chicken/Heyuan/16876/2016 virus and at 8 dpi in one dead mouse challenged with the A/Chicken/Huizhou/HZ-3/2016 virus. The sequences were confirmed by NGS. Polymorphisms of the E627K and D701N mutations in the PB2 gene were identified in the A/Guangdong/Th008/2017-infected mice by NGS. The viruses were in dynamic mutation.
FIG 4.
Challenge of chickens and mice with five H7N9 viruses. The five challenge viruses were A/Chicken/Guangdong/SW154/2015(H7N9), A/Chicken/Heyuan/16876/2016(H7N9), A/Chicken/Huizhou/HZ-3/2016(H7N9), A/Guangdong/Th005/2017(H7N9), and A/Guangdong/Th008/2017(H7N9). (A) Mortality of inoculated chickens. Groups of 10 6-week-old SPF chickens were inoculated intranasally with 105 EID50/200 μl of each virus. (B) Virus titers in the lungs and brains of the first three dead inoculated chickens. (C) Mortality of groups of eight 6-week-old SPF chickens in direct contact with infected chickens. (D) Virus titers in the lungs and brains of the first three dead chickens, infected chickens euthanized at 3 dpi, and naive chickens in direct contact with infected chickens at 4 dpi. (E) Mouse mortality. Four- to 5-week-old female BALB/c mice (13 mice/group) were inoculated intranasally with 106 EID50 of each virus. (F) Virus titers in the lungs and brains of mice at necropsy at 4 dpi. (G) Weight changes in the mice.
Peking ducks challenged with all five H7N9 viruses or those in contact with infected ducks showed no clinical signs, but 20%, 60%, and 20% of the ducks challenged with A/Chicken/Heyuan/16876/2016, A/Guangdong/Th005/2017, and A/Guangdong/Th008/2017, respectively, showed virus shedding at the different sampling time points and seroconversion at 14 dpi. None of the other infected ducks or ducks exposed by direct contact showed virus shedding or seroconversion.
DISCUSSION
The long-term circulation of H7N9 LPAIV has attracted global attention, particularly because of its ability to cause human infections. A key concern is that the continued circulation of H7N9 AIVs in domestic poultry may lead to the emergence of H7N9 HPAIVs, including those capable of infecting humans. During our AIV surveillance, we discovered in mid-2016 an H7N9 HPAIV variant that possessed the insertion of multiple amino acids at the cleavage site of the HA protein. Strikingly, in other H7N9 HPAIVs detected since then, three novel amino acid motifs have been identified, and two of these have recently caused human infections in Guangdong Province (Guangzhou City, Qingyuan City, and Shenzhen City) and Taiwan (12). The insertion of a cleavage site motif with the insertion of multiple basic amino acids has previously been described in AIVs of the H7N7 and H5N3 subtypes via consecutive cultures on chicken embryo cells or repeated passage in chickens (13, 14). However, the mechanism of insertion of amino acids is not clear (15).
Although our data suggest that the original source of the novel H7N9 HPAIVs is a virus of the Yangtze River Delta lineage, it is notable that the novel highly pathogenic H7N9 variants were first reported in the Pearl River Delta region in Guangdong Province, with these isolates being recovered in five cities over an approximately 6-month period. This, combined with the relatively long branch separating the novel H7N9 HPAIV variants and those in wave 3 in the HA and NA phylogenies, raises the possibility that the novel H7N9 HPAIV variants might have originated from a wave 3-like virus that previously circulated at a low level in Guangdong Province. However, because only limited sequence data are available, we cannot exclude the possibility that the novel H7N9 HPAIVs emerged outside Guangdong and were imported into this province in or before July 2016.
The HA and NA gene trees also revealed that some viruses from different geographic regions (e.g., the Guangxi region, Fujian Province, and Hong Kong) belonged to the Pearl River Delta lineage, suggestive of virus transmissions from Guangdong to neighboring provinces (see Fig. S1 and S2 in the supplemental material). Again, however, because only limited sequence data are available, we cannot exclude the possibility that the novel H7N9 HPAIV variants emerged outside Guangdong and were imported into this province in or before July 2016. More recently, nine highly pathogenic H7N9 AIV outbreaks have been reported on chicken farms in eight provinces, including Hunan, Hebei, Henan, Anhui, Shanxi, Heilongjiang, Inner Mongolia, and Tianjin. More seriously, three provinces have reported human infections with H7N9 HPAIV (10). Therefore, although the highly pathogenic H7N9 variants have, to date, largely been observed in Guangdong Province, there is a clear risk that they will spread to other regions (Fig. 2D) through the transportation of live poultry (16, 17). The potential spread and continuing evolution of the novel HPAIV H7N9 variants evidently require additional study.
The 226Q mutation in the HA of the HPAIVs from poultry and humans suggests that these variants preferentially bind to avian-like influenza virus receptors (18, 19, 30), and the S31N mutation of M2 suggests that these variants are resistant to adamantanes (20). Of note, none of the chicken viruses but three of the four human HPAIV isolates had the E627K mutation, two human viruses had an A588V substitution, while two other human viruses carried the K526R mutation; hence, these mutational patterns suggest that these human-origin viruses have adapted to some extent to replicate in mammals (21, 22), and the quick adaptation mutations occurring in mice were confirmed by NGS. In addition, three of the four human HPAIV strains possessed the R292K mutation in NA that is indicative of resistance to neuraminidase inhibitors (NAIs) (23, 24). All the NAI resistance-conferring mutations emerged in human H7N9 LPAIVs, including in the HPAIV A/Guangdong/Th008/2017(H7N9), during treatment (8) (Table S2). The mammal-adapted and NAI resistance-conferring mutations were not found in the H7N9 HPAIV variants of chicken origin, again indicating a quick adaptation mutation in humans.
The existence of multiple basic amino acids at the cleavage site of the HA protein is regarded as a hallmark of high pathogenicity for chickens (25). Our animal experiments showed that these novel H7N9 HPAIV variants are highly pathogenic for chickens and the human-origin viruses were more highly pathogenic than avian-origin viruses in mice. Notably, the human-origin viruses that contained mutations for adaptation to mammals displayed a higher level of virulence in mice than the H7N9 viruses of chicken origin. In addition, the chicken-origin viruses evolved rapidly to contain mutations for adaptation to mammals, which were seen by 4 dpi in mice. Hence, this study reinforces concerns that the continued circulation of H7N9 AIVs in domestic poultry may eventually lead to the emergence of H7N9 HPAIV variants capable of infecting humans. Clearly, the further potential adaptation of this novel highly pathogenic virus in poultry warrants additional investigation and ongoing surveillance in bird and human populations.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were carried out in animal biosafety level 3 (ABSL-3) facilities in compliance with the protocols of the Biosafety Committee of the ABSL-3 Laboratory of the South China Agriculture University (CNAS BL0011). All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the South China Agriculture University and were carried out in accordance with the approved guidelines (2017A002).
RNA extraction, RT-PCR, and DNA sequencing.
RNA was extracted from a suspension of virus isolates with an RNeasy minikit (Qiagen) as directed by the manufacturer. A two-step reverse transcription-PCR (RT-PCR) was conducted with universal primers as reported by Hoffmann et al. (26), and every gene segment was amplified under standard conditions. PCR products were purified with a QIAamp gel extraction kit (Qiagen) and sequenced with an ABI 3730 DNA analyzer (Applied Biosystems).
Real-time RT-PCR.
A real-time reverse transcription-PCR (rRT-PCR) assay was developed for the detection of H7N9 highly pathogenic avian influenza A viruses (HPAIVs) (11). The specific primers and probe sets used for the detection of the HA genes were as follows: forward primer HA976F (5′-GCAACAGGGATGAAGAATGTTCC-3′), reverse primer HA1097R (5′-CCATACCAACCATCAATTAGGCC-3′), and probe HA1011 (5′-FAM-GAGAAAACGGACTGCGAGAGGCC-BHQ1-3′); the probe was labeled with the reporter 6-carboxyfluorescein (FAM) and black hole quencher 1 (BHQ1). The primers and probes were synthesized by Sangon (Shanghai, China). The assay was conducted using an AgPath-ID one-step RT-PCR kit (Life Technologies) on an Applied Biosystems 7500 real-time PCR system with the following conditions: 50°C for 10 min, followed by 95°C for 10 min and then 40 cycles of 95°C for 15 s and 60°C for 45 s. Fluorescence was recorded at 60°C.
Viruses used for animal experiments.
A/Chicken/Guangdong/SW154/2015(H7N9), A/Chicken/Heyuan/16876/2016(H7N9), and A/Chicken/Huizhou/HZ-3/2016(H7N9) were isolated from LPMs in Guangdong Province, China. Two human H7N9 viruses, A/Guangdong/Th005/2017(H7N9) and A/Guangdong/Th008/2017(H7N9), were isolated from two patients in Guangdong Province from late 2016 to early 2017 (8). Viruses were propagated in 9- to 11-day-old specific-pathogen-free (SPF) embryonated chicken eggs and stored at −70°C until use.
Chicken experiments.
In the first chicken experiment, the intravenous pathogenicity index (IVPI) test was performed to determine the pathogenicity of the five H7N9 viruses according to the recommendations of the World Organisation for Animal Health (OIE) (27). In brief, 10 6-week-old SPF chickens were intravenously inoculated with 0.1 ml of a 1/10 dilution of fresh infectious allantoic fluid, and two 6-week-old SPF chickens were inoculated with 0.1 ml of phosphate-buffered saline and used as a control group. All chickens were examined daily for 10 days. At each observation, each chicken was scored on the basis of its condition, as follows: 0 for normal, 1 for sick, 2 for severely sick, and 3 for dead. IVPI was the mean score per chicken per observation over the 10-day period.
In a second chicken experiment, groups of 10 6-week-old SPF chickens (Jinan Sipafas Poultry Co., Ltd., Shandong Province, China) were inoculated intranasally with 105 EID50/200 μl of the five H7N9 viruses. Thirteen chickens were used as negative controls. Chickens were observed for clinical symptoms for 14 days, and serum samples were collected for analysis of virus-specific antibodies. Eight naive sentinel chickens were introduced into the isolators with infected chickens 24 h after the latter chickens were directly inoculated to assess transmission. Cloacal and throat swab specimens were collected at 3, 5, 7, 9, 11, and 13 dpi from the infected and naive chickens. The first three dead chickens in each group were dissected to test for virus replication in the organs, including the heart, lung, kidney, brain, spleen, and liver. The three chickens in the group infected with LPAIV A/Chicken/Guangdong/SW154/2015 and the naive contact group were euthanized on 4 dpi to test for virus in the organs. Tissue and swab samples were collected for virus titration by an EID50 assay.
Duck experiments.
Ten 4-week-old influenza virus-seronegative Peking ducks were inoculated intranasally with 106 EID50/200 μl of the five H7N9 viruses. Ten ducks were used as negative controls. All of the ducks were observed for clinical signs for 14 days. At 1 dpi, eight naive sentinel ducks were introduced into the isolators with the infected ducks to assess transmission. Cloacal and throat swab specimens were collected from the infected ducks and the naive ducks at 3, 5, 7, 10, and 14 dpi. The ducks were humanely euthanized at 14 dpi, and serum samples were collected for analysis of virus-specific antibodies.
Mouse experiments.
Groups of 13 4- to 5-week-old female BALB/c mice (weight, 14 to 16 g), obtained from the Guangdong Medical Lab Animal Center, were anesthetized with CO2 and inoculated intranasally with 106 EID50/50 μl of the five H7N9 viruses indicated above. Body weight and clinical signs were monitored daily for 14 days after infection. The mice were euthanized if they lost more than 25% of their initial body weight. To test for virus replication in the organs, three mice in each group were euthanized at 4 dpi and tissue samples, including brain and lung tissue samples, were collected for virus titration by an EID50 assay. Serum samples were collected for analysis of virus-specific antibodies at 14 dpi.
To investigate the adaptation of the two avian H7N9 viruses in the mammal model, groups of 10 3- to 4-week-old female BALB/c mice (weight, 12 to 14 g) were inoculated intranasally with 106 EID50/50 μl of the A/Chicken/Heyuan/16876/2016 or A/Chicken/Huizhou/HZ-3/2016 virus. Three mice in each group were euthanized at 4 dpi, and lung tissue samples were collected for Sanger sequencing of the viruses. The lungs of dead mice were subjected to the same procedure for Sanger sequencing of the viruses.
Phylogenetic analysis.
To determine the phylogenetic position of the newly isolated strains in our study, we performed phylogenetic analyses of the HA and NA genes. All of the available HA and NA genes with the complete coding region of H7N9 were downloaded from GenBank and GISAID and combined with our data. Two sequence data sets were then created: the HA gene (n = 976) and NA gene (n = 985) data sets (sequence alignments are available on request). All sequences were aligned using the MAFFT (version 7.149) program (28). Maximum likelihood (ML) phylogenies for the codon alignment of each gene segment were estimated using the GTRGAMMA nucleotide substitution model in the RAxML (version 8.2) program (29). Node support was determined by nonparametric bootstrapping with 1,000 replicates. All phylogenetic trees were rooted on the earliest sampled virus (A/Shanghai/1/2013) and visualized in the FigTree (version 1.4.3) program (http://tree.bio.ed.ac.uk/software/figtree/).
Next-generation sequencing.
The five challenge virus strains and viruses from lung samples from infected mice and chickens were sequenced using next-generation sequencing (NGS) on an Illumina HiSeq 2000 sequencer by BGI-Shenzhen.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the authors and originating laboratories submitting the sequences to GISAID's EpiFlu Database on which this research is based, especially the WHO Chinese National Influenza Center Virology Institute, the Chinese CDC, and the Guangzhou Institute of Respiratory Diseases. All submitters of data may be contacted directly via the GISAID website (www.gisaid.org). We also acknowledge the contributions of Tao Jin and Guang Liu of BGI-Shenzhen for NGS analysis.
This work was partially supported by the National Natural Science Foundation (NSFC) of China (31672586, U1501212, 31471253, 81470096), the Science and Technology Planning Project of Guangdong Province (2013B020224), and the National Key Research and Development Program of China (2016YFC1200800, 2016YFD0500201 and 2016YFD0500800). W. Qi and M. Liao are supported by the Special Support Plan of Guangdong Province in Science and Technology for Talents. G. F. Gao is a leading principal investigator of the NSFC Innovative Research Group (81621091). W. Shi is supported by the Taishan Scholar project of Shandong Province. J. Cui is supported by the CAS Pioneer Hundred Talents Program. D. Liu is supported by the National Program for Support of Top-Notch Young Professionals. E. C. Holmes is supported by an NHMRC Australia fellowship (GNT1037231). Y. Bi is supported by the Youth Innovation Promotion Association of the Chinese Academy of Sciences (CAS) (2017122).
W. Qi, W. Jia, D. Liu, Y. Bi, J. Cui, J. Li, W. Shi, E. C. Holmes, G. Zhang, T. Ren, G. F. Gao, and M. Liao conceived and designed the experiments. W. Qi, W. Jia, S. Xie, B. Li, L. Xing, J. Zhang, H. Li, G. Su, G. Lao, C. Xu, and M. Liao performed the experiments. W. Qi, W. Jia, D. Liu, J. Li, Y. Bi, T. Hu, B. Li, Y. Du, X. Wei, J.-S. Eden, H. Tian, W. Li, B. Xu, W. Liu, J. Cui, F. Zhang, W. Shi, E. C. Holmes, G. F. Gao, and M. Liao analyzed the results of the experiments. W. Qi, W. Jia, D. Liu, Y. Bi, J. Cui, E. C. Homes, W. Shi, F. Zhang, J.-S. Eden, B. Li, G. Zhang, T. Ren, G. F. Gao, and M. Liao wrote and modified the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00921-17.
REFERENCES
- 1.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 2.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam TT, Zhou B, Wang J, Chai Y, Shen Y, Chen X, Ma C, Hong W, Chen Y, Zhang Y, Duan L, Chen P, Jiang J, Zhang Y, Li L, Poon LL, Webby RJ, Smith DK, Leung GM, Peiris JS, Holmes EC, Guan Y, Zhu H. 2015. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature 522:102–105. doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, Yang L, Zhu W, Zhang Y, Zou S, Bo H, Gao R, Dong J, Huang W, Guo J, Li Z, Zhao X, Li X, Xin L, Zhou J, Chen T, Dong L, Wei H, Li X, Liu L, Tang J, Lan Y, Yang J, Shu Y. 2016. Two outbreak sources of influenza A H7N9 viruses have been established in China. J Virol 90:5561–5573. doi: 10.1128/JVI.03173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou L, Ren R, Yang L, Bao C, Wu J, Wang D, Li C, Xiang N, Wang Y, Li D, Sui H, Shu Y, Feng Z, Li Q, Ni D. 2017. Sudden increase in human infection with avian influenza A H7N9 virus in China, September-December 2016. Western Pac Surveill Response J 8:1. doi: 10.5365/WPSAR.2017.8.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Monthly risk assessment summary. World Health Organization, Geneva, Switzerland: http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/. [Google Scholar]
- 7.Cui L, Liu D, Shi W, Pan J, Qi X, Li X, Guo X, Zhou M, Li W, Li J, Haywood J, Xiao H, Yu X, Pu X, Wu Y, Yu H, Zhao K, Zhu Y, Wu B, Jin T, Shi Z, Tang F, Zhu F, Sun Q, Wu L, Yang R, Yan J, Lei F, Zhu B, Liu W, Ma J, Wang H, Gao GF. 2014. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 5:3142. doi: 10.1038/ncomms4142. [DOI] [PubMed] [Google Scholar]
- 8.Zhang F, Bi Y, Wang J, Wong G, Shi W, Hu F, Yang Y, Yang L, Deng X, Jiang S, He X, Liu Y, Yin C, Zhong N, Gao GF. 2017. Human infections with recently-emerging highly pathogenic H7N9 avian influenza in China. J Infect 75:71–75. doi: 10.1016/j.jinf.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, Tan Y, Kang M, Liu F, Ren R, Wang Y, Chen T, Yang Y, Li C, Wu J, Zhang H, Li D, Greene CM, Zhou S, Iuliano AD, Havers F, Ni D, Wang D, Feng Z, Uyeki TM, Li Q. 2017. Preliminary epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus. Emerg Infect Dis 23:1355–1359. doi: 10.3201/eid2308.170640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Organisation for Animal Health. 2017. Follow-up report no. 6. World Organisation for Animal Health, Paris, France. http://www.oie.int/wahis_2/temp/reports/en_fup_0000024088_20170623_122928.pdf.
- 11.Jia W, Cao C, Lin Y, Zhong L, Xie S, Wang X, Yin S, Xu Z, Dai Y, Li Z, Niu X, Qi W, Lu T, Liao M. 2017. Detection of a novel highly pathogenic H7 influenza virus by duplex real-time reverse transcription polymerase chain reaction. J Virol Methods 246:100–103. doi: 10.1016/j.jviromet.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Yang JR, Liu MT. 2017. Human infection caused by an avian influenza A H7N9 virus with a polybasic cleavage site in Taiwan, 2017. J Formos Med Assoc 116:210–212. doi: 10.1016/j.jfma.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Li SQ, Orlich M, Rott R. 1990. Generation of seal influenza virus variants pathogenic for chickens, because of hemagglutinin cleavage site changes. J Virol 64:3297–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito T, Goto H, Yamamoto E, Tanaka H, Takeuchi M, Kuwayama M, Kawaoka Y, Otsuki K. 2001. Generation of a highly pathogenic avian influenza A virus from an avirulent field isolate by passaging in chickens. J Virol 75:4439–4443. doi: 10.1128/JVI.75.9.4439-4443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suarez DL. 2017. Influenza A virus, p 3–30. In Swayne DE. (ed), Animal influenza. Wiley-Blackwell, Ames, IA. [Google Scholar]
- 16.Gao GF. 2014. Influenza and the live poultry trade. Science 344:235. doi: 10.1126/science.1254664. [DOI] [PubMed] [Google Scholar]
- 17.Su S, Bi Y, Wong G, Gray GC, Gao GF, Li S. 2015. Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J Virol 8917:8671–8676. doi: 10.1128/JVI.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Xiao H, Wu Y, Liu D, Qi X, Shi Y, Gao GF. 2014. H7N9: a low pathogenic avian influenza A virus infecting humans. Curr Opin Virol 5:91–97. doi: 10.1016/j.coviro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, Qin C, Sun H, Liu J, Haywood J, Liu W, Gong W, Wang D, Shu Y, Wang Y, Yan J, Gao GF. 2013. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- 20.Hay AJ, Zambon MC, Wolstenholme AJ, Skehel JJ, Smith MH. 1986. Molecular basis of resistance of influenza A viruses to amantadine. J Antimicrob Chemother 18:19–29. doi: 10.1093/jac/18.Supplement_B.19. [DOI] [PubMed] [Google Scholar]
- 21.Xiao C, Ma W, Sun N, Huang L, Li Y, Zeng Z, Wen Y, Zhang Z, Li H, Li Q, Yu Y, Zheng Y, Liu S, Hu P, Zhang X, Ning Z, Qi W, Liao M. 2016. PB2-588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci Rep 6:19474. doi: 10.1038/srep19474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi Y, Xie Q, Zhang S, Li Y, Xiao H, Jin T, Zheng W, Li J, Jia X, Sun L, Liu J, Qin C, Gao GF, Liu W. 2015. Assessment of the internal genes of influenza A (H7N9) virus contributing to high pathogenicity in mice. J Virol 89:2–13. doi: 10.1128/JVI.02390-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Bi Y, Vavricka CJ, Sun X, Zhang Y, Gao F, Zhao M, Xiao H, Qin C, He J, Liu W, Yan J, Qi J, Gao GF. 2013. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res 23:1347–1355. doi: 10.1038/cr.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke C, Mok CKP, Zhu W, Zhou H, He J, Guan W, Wu J, Song W, Wang D, Liu J, Lin Q, Chu DKW, Yang L, Zhong N, Yang Z, Shu Y, Peiris JSM. 2017. Human infection with highly pathogenic avian influenza A(H7N9) virus, China. Emerg Infect Dis 23:1332–1340. doi: 10.3201/eid2308.170600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409–417. doi: 10.1016/S0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 14612:2275–2289. [DOI] [PubMed] [Google Scholar]
- 27.World Organisation for Animal Health. 2017. Avian influenza (infection with avian influenza viruses). In OIE manual of diagnostic tests and vaccines for terrestrial animals. World Organisation for Animal Health, Paris, France. [Google Scholar]
- 28.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Shi Y, Zhang W, Zhang Y, Gao GF. 2013. Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut). Protein Cell 4:502–511. doi: 10.1007/s13238-013-3906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.