Figure 5. Sde2 is required for excision of selected introns from a subset of pre‐mRNAs.
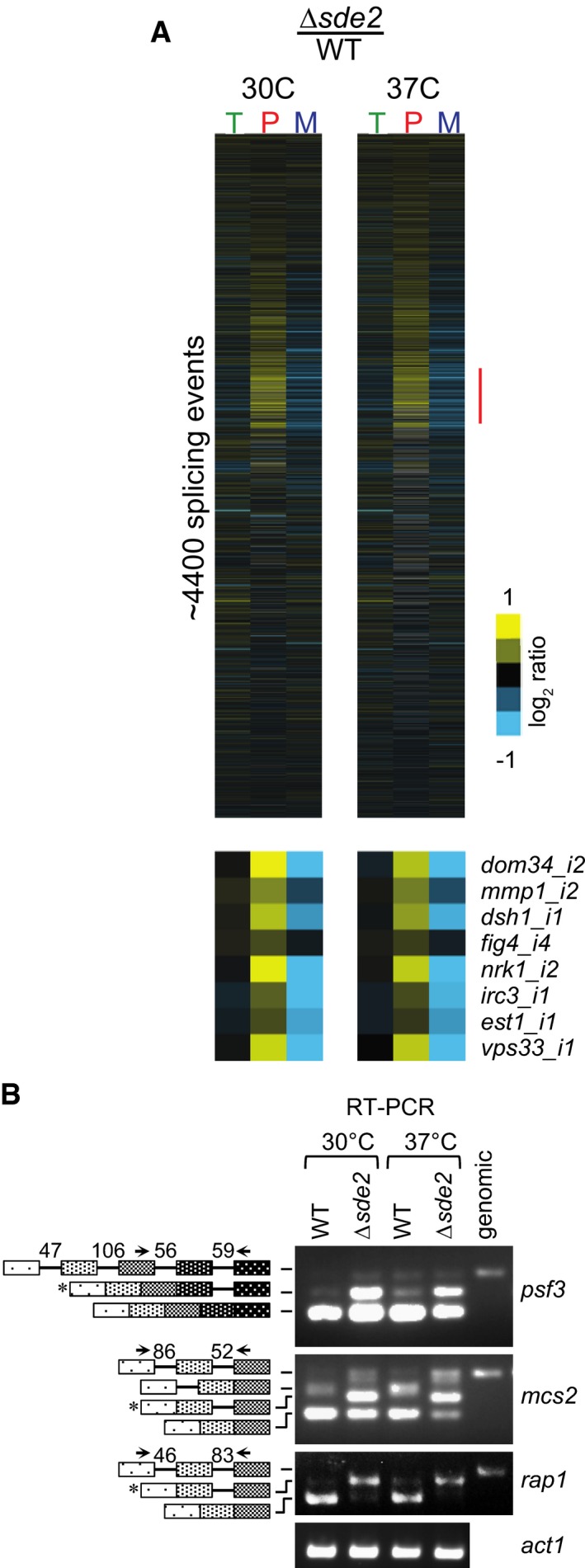
- Analysis of total RNA from Schizosaccharomyces pombe WT and Δsde2 strain using splicing‐sensitive microarray. Microarray heat‐map shows log2 Δsde2/WT ratio of signals for total transcripts (T), intron‐containing transcripts (P) and spliced transcripts (M). 37°C temperature shift was performed for 15 min to monitor early splicing defects. The experiment was repeated with dyes swapped. Yellow colour represents accumulation, black denotes no change, and blue shows reduction of signal in Δsde2.
- Semi‐quantitative RT–PCR shows intron‐containing transcripts for psf3, mcs2 and rap1. cDNA prepared from total RNA isolated from S. pombe WT and Δsde2 mutant at 30°C and 37°C was analysed by PCR. The block diagram (not drawn to the scale) shows exons and introns. Arrows mark primers used for amplification. The numbers above introns (shown as lines in block diagrams) mark their lengths. PCR band from genomic DNA (genomic) corresponds to the pre‐mRNA. RT–PCR of act1 (actin) pre‐mRNA is used as control. Asterisks mark intron‐containing transcripts getting enriched in Δsde2.
Source data are available online for this figure.
