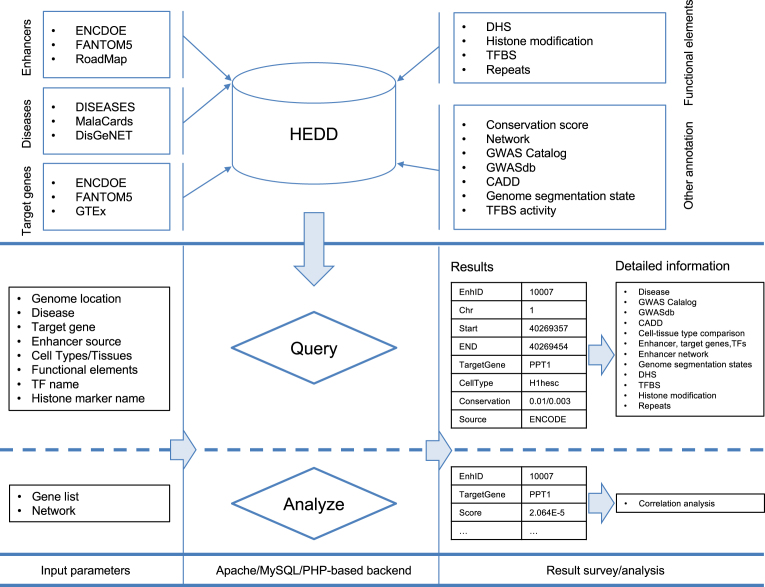
Figure 1.
Database content and construction. HEDD collected the enhancers (from three major epigenome study projects), enhancer target gene and gene disease set to quantify the connections between enhancer and diseases. Besides the disease information, it also stores the genetic and epigenetic information (e.g. DHS, TFBS, conservation score) related to enhancers. Users can query with multiple options (e.g. genome locations, disease name, gene name) to acquire enhancers and further view all the detail information such as, associated disease, cell type/tissue, overlapped GWAS SNPs, CADD score and neighboring enhancer for a specific enhancer. It enables users to do scoring analysis: for a gene set of interest, users can score enhancers in a gene network for their ‘relatedness’ to the gene set. All results of query and analysis can be downloaded for further analysis. DHS: DNase I hypersensitive sites, GWAS: genome-wide association studies, TF: transcription factor, TFBS: transcription factor biding sites, CADD: combined annotation dependent depletion.