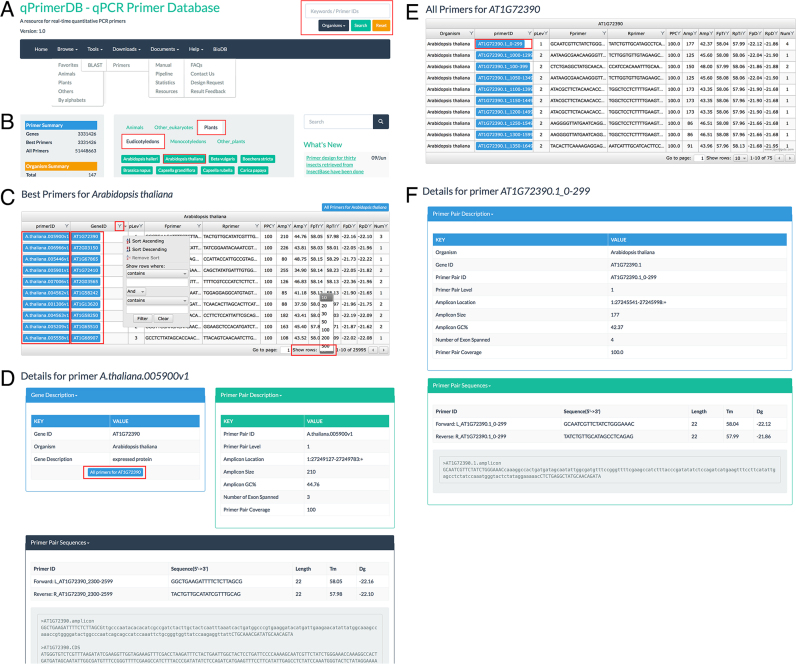
Figure 2.
Screenshots of the navigation bar and browse module in qPrimerDB. (A) Navigation bar of qPrimerDB. A search box is provided in the upper-right corner of every page to enable convenient searching of keywords of interest. (B) Browse interface on the ‘Home’ page. Users can browse the organism of interest, for example, by sequentially clicking ‘Plants’, ‘Eudicotyledons’ and ‘Arabidopsis thaliana’. (C) Example of A. thaliana primers in table format. Both the record number per page and the order of each column can be adjusted, as needed. (D) qPrimerDB primer details page. Detailed information for primer ID A. thaliana.005900.v1 is presented in three sections: Gene Description (gene ID, organism, gene description, and a blue button ‘All primers for AT1G72390’); Primer Pair Description (primer pair ID and level, amplicon location, amplicon size, amplicon GC content, number of exons spanned, PPC); and Primer Pair Sequences (Tm values of primers, primer sequences, primer length and amplicon and template coding/mRNA sequences). (E) Example of all primers for AT1G72390 in table format. After clicking the blue button in Figure 2D, all primers will be listed in table format. (F) Detailed information for primer ID AT1G72390.1_0–299 is presented in two sections: Primer Pair Description (organism, Gene ID, primer pair ID and level, amplicon location, amplicon size, amplicon GC content, number of exons spanned, PPC); and Primer Pair Sequences (Tm values of primers, primer sequences, primer length and amplicon sequence).