Abstract
We present RISE (http://rise.zhanglab.net), a database of RNA Interactome from Sequencing Experiments. RNA-RNA interactions (RRIs) are essential for RNA regulation and function. RISE provides a comprehensive collection of RRIs that mainly come from recent transcriptome-wide sequencing-based experiments like PARIS, SPLASH, LIGR-seq, and MARIO, as well as targeted studies like RIA-seq, RAP-RNA and CLASH. It also includes interactions aggregated from other primary databases and publications. The RISE database currently contains 328,811 RNA-RNA interactions mainly in human, mouse and yeast. While most existing RNA databases mainly contain interactions of miRNA targeting, notably, more than half of the RRIs in RISE are among mRNA and long non-coding RNAs. We compared different RRI datasets in RISE and found limited overlaps in interactions resolved by different techniques and in different cell lines. It may suggest technology preference and also dynamic natures of RRIs. We also analyzed the basic features of the human and mouse RRI networks and found that they tend to be scale-free, small-world, hierarchical and modular. The analysis may nominate important RNAs or RRIs for further investigation. Finally, RISE provides a Circos plot and several table views for integrative visualization, with extensive molecular and functional annotations to facilitate exploration of biological functions for any RRI of interest.
INTRODUCTION
RNA molecules in the cell do not exist alone. During their life cycle, they interact with many different molecules, including proteins, DNA and other RNAs (1–4). These interactions are essential to understanding the biological functions and molecular mechanisms of both messenger RNAs (mRNAs) and noncoding RNAs (ncRNAs). RNA molecules can directly interact with other RNAs through base-pairing. For instance, the 3′ UTRs of mRNAs can be targeted by miRNAs, and the intronic regions of pre-mRNAs can be recognized by the spliceosomal small nuclear RNAs (snRNAs). In addition to protein-coding mRNAs and canonical ncRNAs, mammalian genomes contain many thousands of long noncoding RNAs (lncRNAs) (5–7). Some lncRNAs play important and diverse roles in gene regulation through interactions with other RNAs (8–10). For example, many lncRNAs can be competing targets of shared miRNAs with other mRNAs, forming a complex regulatory competing endogenous RNA (ceRNA) network (3,11). These observations indicate that intermolecular RNA-RNA interactions (RRIs) may be a general strategy used by RNA molecules in the cell. Collection of RRIs thus may provide insights into the biological functions and regulatory mechanisms of both mRNAs and ncRNAs.
Mapping in vivo RRIs had remained challenging until the recent development of several sequencing-based technologies. For example, CLASH (12,13), hiCLIP (14), RIA-seq (15) and RAP-RNA (16) can detect RRIs for a target RNA or a protein. More recently, some techniques have been developed to identify transcriptome-wide RRI networks (i.e. RNA interactomes). For example, PARIS (17), SPLASH (18) and LIGR-seq (19) can massively discover direct RRIs in a cell; and MARIO (20) can map RRIs assisted by proteins.
The RRIs generated by these large-scale studies have not been systematically collected and analyzed. Currently, there are several databases that contain RRI information, such as NPInter (21), RAID (22) and RAIN (23). But these databases focus mainly on miRNAs-mRNA interactions (Supplementary Table S1), i.e. RRIs of miRNA targeting. In addition, RRIs in these databases usually contain little information about their cell types, resolving technologies, etc. However, RRIs are highly dynamic in vivo (17); thus it will be important to include these details in annotation and analysis. Furthermore, RRIs in these databases are often of limited resolution and do not contain precise interacting sites on the RNA transcripts.
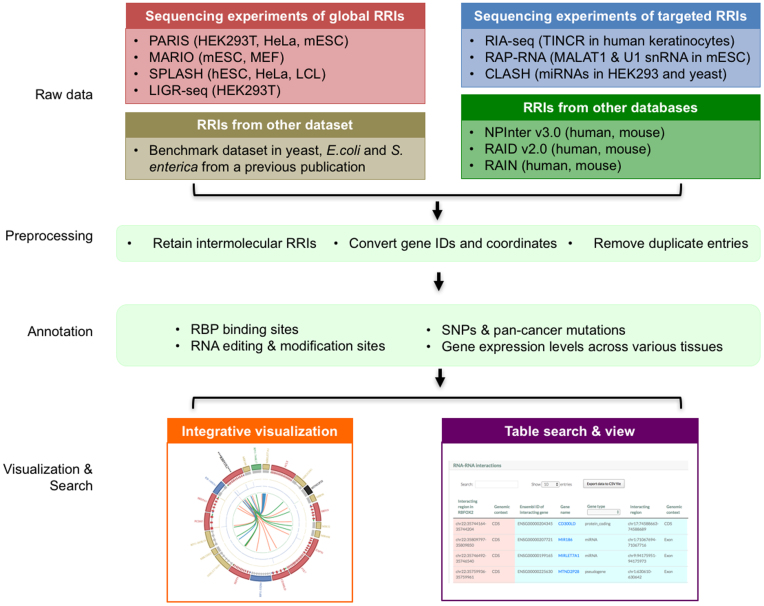
To address these challenges, we construct RISE, a comprehensive database of RNA Interactome from Sequencing Experiments (Figure 1). The RRIs are from recently developed transcriptome-wide and targeted sequencing-based experiments, as well as several primary databases and publications (Table 1). Based on the RISE database, we are then able to compare different RRI datasets and study the network characteristics of the global RNA interactomes. RISE also annotates each RRI with extensive molecular and functional information. The database is a ready-to-use resource for researchers looking for interaction and other functional information on individual RNAs, and analyzing RRI networks of specific pathways or systems.
Figure 1.
Framework to construct the RISE database. We collected RRIs from transcriptome-wide and targeted sequencing experiments, and other databases and publications. We performed quality control to obtain non-redundant intermolecular RRI entries. We then annotated RRIs with extensive molecular and functional information, including (i) RBP binding sites, (ii) RNA editing and modification sites, (iii) SNPs and pan-cancer mutations, and (iv) gene expression levels from various cell and tissue types. Finally, RISE provides integrative Circos plot visualization and table views for the search results.
Table 1. Overview of data collected in the RISE database.
| Category | Method/Resource | Species | Cell line | Number of interactions | Number of involved genes |
|---|---|---|---|---|---|
| Transcriptome-wide studies | PARIS | Human | HEK293T | 25 824 | 16 192 |
| Human | Hela (high RNase) | 25 552 | 19 335 | ||
| Human | Hela (low RNase) | 20 330 | 17 009 | ||
| Mouse | mESC | 29 514 | 12 625 | ||
| MARIO | Mouse | MEF | 7 167 | 2 936 | |
| Mouse | mESCa | 99 290 | 15 309 | ||
| Mouse | mESCb | 37 441 | 9 715 | ||
| SPLASH | Human | hESC | 3 345 | 971 | |
| Human | HeLa | 5 799 | 1 649 | ||
| Human | LCL | 4 213 | 429 | ||
| Human | hESC (RA treated) | 1 770 | 671 | ||
| LIGR-seq | Human | HEK293T | 641 | 749 | |
| Targeted studies | RIA-seq | Human | Keratinocytes (TINCR) | 3 609 | 1 815 |
| RAP-RNA | Mouse | mESC (Malat1) | 495 | 489 | |
| Mouse | mESC (U1 snRNA) | 12 278 | 8 635 | ||
| CLASH | Human | HEK293 (miRNAs) | 18 508 | 7260 | |
| Yeast | BY4741 (miRNAs) | 253 | 47 | ||
| From other databases/dataset | NPInter v3.0 | Human | − | 3 691 | 2 525 |
| Mouse | − | 52 | 83 | ||
| RAID v2.0 | Human | − | 22 521 | 6 262 | |
| Mouse | − | 3 440 | 2 130 | ||
| RAIN | Human | − | 2 881 | 1 189 | |
| Mouse | − | 36 | 31 | ||
| PMID 26673718 | E. coli | − | 64 | 68 | |
| S. enterica | − | 45 | 49 | ||
| Yeast | − | 52 | 45 | ||
| Total | − | − | − | 328 811 | 56 295 |
aThis experiment uses UV-crosslinking to detect RRIs mediated by one protein.
bThis experiment uses chemical crosslinking to detect RRIs mediated by multiple proteins.
DATA COLLECTION AND ANALYSIS
RRIs from sequencing-based experiments
The RRIs in RISE mainly come from sequencing-based experiments, including global (i.e. transcriptome-wide) studies like (i) PARIS, (ii) SPLASH, (iii) MARIO and (iv) LIGR-seq, as well as targeted studies like (v) RIA-seq, (vi) RAP-RNA and (vii) CLASH. Notably, hiCLIP dataset was not included in RISE because it only provides RRIs within the same RNA transcript (i.e. intramolecular RRIs) (14). Among the global studies, we processed the raw data and identified the RRIs for PARIS because the processed data was not directly available (17); while for the other technologies, we obtained the RRIs from the correspondent publications.
(i) For PARIS, we identified RNA duplex regions using the computational method in the reference (17). Briefly, we first downloaded the raw data from GSE74353, and mapped reads to transcriptomes using STAR (24). We took only the longest isoform as the representative transcript if a gene has multiple transcripts. Then we used in-house scripts (https://github.com/qczhang/paris) to identify RNA duplex regions following the protocols in the reference (17). Finally, only intermolecular RRIs were retained.
(ii, iii) For SPLASH and MARIO, we downloaded the processed data from https://csb5.github.io/splash and http://mariotools.ucsd.edu/legacy/Data_Resources.html, respectively. (iv–vii) For LIGR-seq, RIA-seq, RAP-RNA and CLASH, we obtained the processed data from the references (19), (15), (16) and (12,13) respectively.
RRIs from other databases
RISE also includes RRIs curated from other primary databases and publications, including (i) RAIN, (ii) RAID v2.0, (iii) NPInter v3.0 and (iv) a dataset of experimentally confirmed RRIs.
(i) For RAIN, we downloaded the dataset from: http://rth.dk/resources/rain. (ii) For RAID v2.0, we downloaded the dataset from: http://www.rna-society.org/raid, and then filtered the entries of non-RRIs or RRIs with confident scores below 0.6. (iii) For NPInter v3.0, we downloaded the dataset from: http://www.bioinfo.org/NPInter, and then selected the set of high-confidence RRIs, which were defined as those from literature mining and supported by low-throughput experiments. (iv) In a comparative study of RRI prediction methods, the authors compiled a dataset of experimentally confirmed snoRNA–rRNA and sRNA–mRNA interactions. We obtained the dataset directly from the reference (25).
Comparison and analysis of RRIs
For all datasets, to facilitate cross-experiments comparison, we converted the genomic coordinates into hg38 for human and mm10 for mouse using CrossMap (26), when necessary. We also converted the RefSeq IDs into Ensembl gene IDs using BioMart (27), and the miRNA names into Ensembl gene IDs using miRBase (28). We only retained intermolecular RRIs in all these datasets.
To analyze the RRI networks (i.e. RNA interactomes) in RISE, we first defined a set of unique RRIs in human and mouse by collapsing redundant RRI entries. Finally, the human RNA interactome comprises 112 444 unique RRIs among 29 875 RNA transcripts; while the mouse RNA interactome comprises 166 183 unique RRIs among 22 630 RNA transcripts. Then we used the Python package NetworkX 1.10 (29) to calculate six characteristics of the RNA interactome networks, including (i) distribution of nodes degree, P(k), for nodes of degree k; (ii) distribution of shortest path, S(l), for the shortest paths of length l; (iii) average clustering coefficient, C(k); (iv) average neighborhood connectivity, N(k); (v) average betweenness centrality, B(k) and (vi) average closeness centrality, L(k); all for nodes of degree k.
Molecular annotation for RRIs
To facilitate in-depth investigations of RNA functions and regulations by RISE users, we annotated the genes and interacting regions involved in the RRIs with an array of molecular details. We retrieved RBP binding sites from CLIPdb (30), RNA editing sites from RADAR (31) and DARNED (32), RNA modification sites from RMBase (33), single nucleotide polymorphisms (SNPs) from dbSNP version 142 (34), pan-cancer somatic mutations (35) and gene expression levels in various cell and tissue types from recent publications (36). Finally, the integrative visualization of the RRIs was implemented using a Circos plot (37) and a set of table views.
Database architecture
All metadata in RISE are stored in a MySQL database. The web interface of RISE was implemented with Hyper Text Markup Language (HTML), Cascading Style Sheets (CSS) and Hypertext Preprocessor (PHP). Web design was based on the free templates of Bootstrap (http://getbootstrap.com).
RESULTS AND DATABASE USAGE
Comparing RRIs from sequencing-based experiments
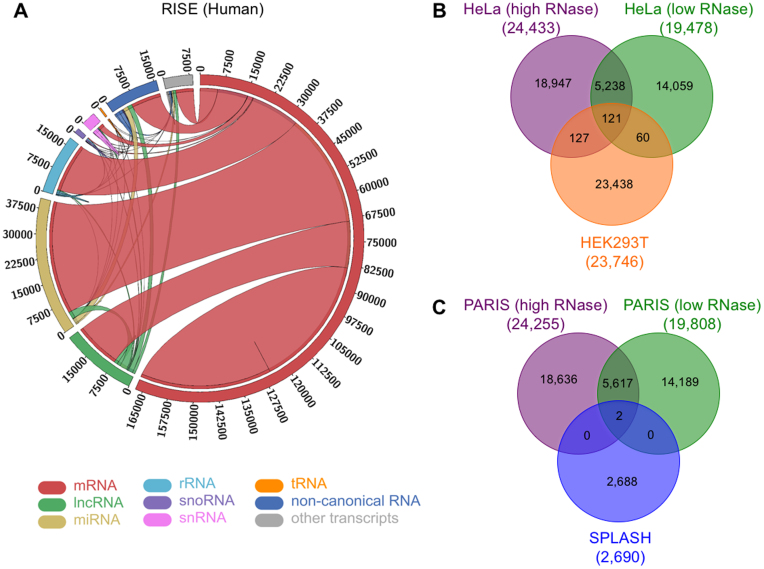
The compilation of a comprehensive RRI repository provides an opportunity to explore RRIs between different types of RNAs, compare RRI datasets from different experiments, and characterize the networks of RNA interactomes. We first analyzed the distribution of RRIs by different RNA types (Figure 2A). We found that ∼90% of the RRIs in human involve mRNAs, in which a substantial fraction is between two different mRNAs. We see a similar distribution in mouse except that there are more interactions with snoRNAs (Supplementary Figure S1). The RRIs involving lncRNAs and miRNAs comprise ∼15% and ∼29% of the total interactions, respectively. In contrast to mRNAs, most of the RRIs involving ncRNAs are formed between ncRNAs and mRNAs. Next, we explored the distributions of RRIs from different experimental approaches and found big cross-platform differences. For example, PARIS detects many RRIs of mRNAs and lncRNAs, while LIGR-seq shows preference on RRIs involving snRNAs, and SPLASH favors RRIs between mRNAs and rRNAs (Supplementary Figure S1).
Figure 2.
Distribution of RRIs by RNA types and comparison of RRIs from different studies in the RISE database. (A) Circos plot showing RRIs between different types of interacting RNAs in human. (B) Overlap of RRIs in different cell lines and experimental conditions detected by the PARIS method. (C) Overlap of RRIs detected by the PARIS and the SPLASH methods in the HeLa cell line.The comparison in (B, C) is counted on the RNA molecular level regardless of the precise interacting regions. And we used the same transcriptome from the SPLASH study as the mapping reference of PARIS in (C) for cross-technology comparison (which explains the RRI number differences between B and C).
The observations above suggest great heterogeneity in the RRI datasets from different experiments. We then systematically analyzed the overlaps among different RRI datasets. We found substantial overlap of RRIs for the same cell line and technology but slightly different experimental specifications (i.e. PARIS with high and low RNase, Figure 2B). The overlap for the same technology but different cell lines are lower although still quite statistically significant. The result suggests the dynamic and cell-specific nature of RNA interactions. But the overlaps between different techniques are very limited even for the same cell line (Figure 2C). One reason is that different techniques have different preferences or biases towards the interactions they can identify. It is also very likely that the RRIs identified by any single experimental approach are far from saturation. One usual drawback of sequencing-based techniques is their heavy dependency on sequencing depth. With low sequencing depth, the stochastic variation may be high.
Characteristics of the RNA interactomes
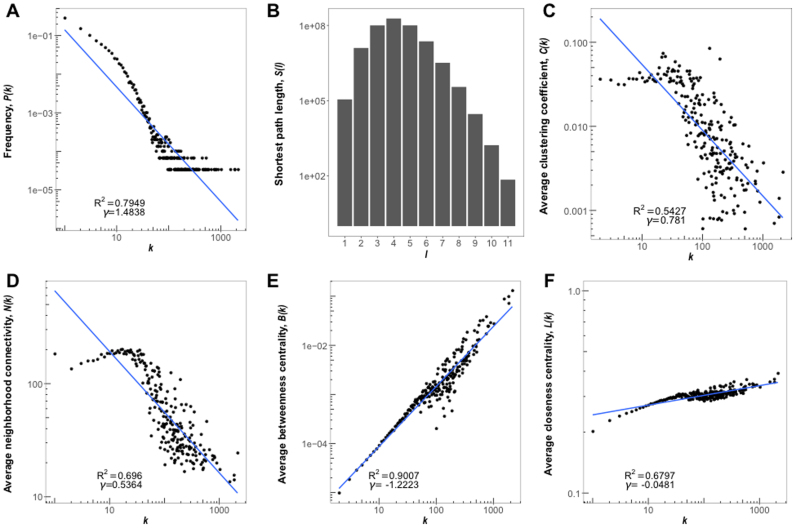
The architectures of global protein–protein interaction networks (i.e. interactomes) have been extensively studied; and a recent study investigated the network features of the RNA-RNA interactions in yeast (38–40). We are also interested in the structural and topological features of the RNA interactomes in RISE. We first calculated the degree distribution P(k) of all RNAs, representing that a given transcript interacts with k other ones. On average, RNAs in the human RRI network have 7 interaction partners, but it also reveals 170 hub RNAs with >100 partners. As shown in Figure 3A, the degree distribution of the human RNAs decreases slowly in a power-law fashion (y∝x−γ). This suggests that, similar to protein interactomes, the human RNA interactome also tends to be scale-free. We then calculate the shortest path length distribution in the largest connected subgraph, S(l), where l means the number of edges in the shortest path between any two nodes. We found that most of them are less than 8 edges, with the longest one to be 11 edges (i.e. network diameter. Figure 3B). This means that most RNAs are very closely linked, suggesting that the human RNA interactome is also a small world network.
Figure 3.
Characteristics of the human RNA interactome in RISE. (A) Degree distribution of the RNAs. (B) Distribution of the shortest path between pairs of RNAs. (C) Degree distribution of the average clustering coefficients of the RNAs. (D) Degree distribution of the average neighborhood connectivity. (E) Degree distribution of the average betweenness centrality of the RNAs. (F) Degree distribution of the average closeness centrality of the RNAs. The blue lines show the regression in log-space.
We also calculated the average clustering coefficient, C(k), and the average neighborhood connectivity, N(k) for all RNAs of the same interaction degree k. Both of them are measures of the tendency of molecules in a network to form local clusters. We found that C(k) and N(k) diminishes when the number of interactions per RNA increases (Figure 3C and D). This suggests that RNAs with low interaction degrees tend to form clusters, i.e. modules. These modules are then connected by RNAs of high interaction degrees, i.e. hubs. It thus indicates a potential topology of hierarchical and modular organization.
We further calculated the average centrality measured by betweenness centrality, B(k), and closeness centrality, L(k) for RNAs of interaction degree k. As shown in Figure 3E and F, interaction degrees and the centralities are positively correlated. This implies that RNA interaction hubs are usually of high centrality in the network and deserve more attention in functional investigation.
We repeated the analysis for mouse interactome and confirmed the finding that, like protein interactomes, RNA interactomes also tend to be scale-free, small-world, hierarchical and modular. And interaction hubs tend to be essential nodes in the networks (Supplementary Figure S2).
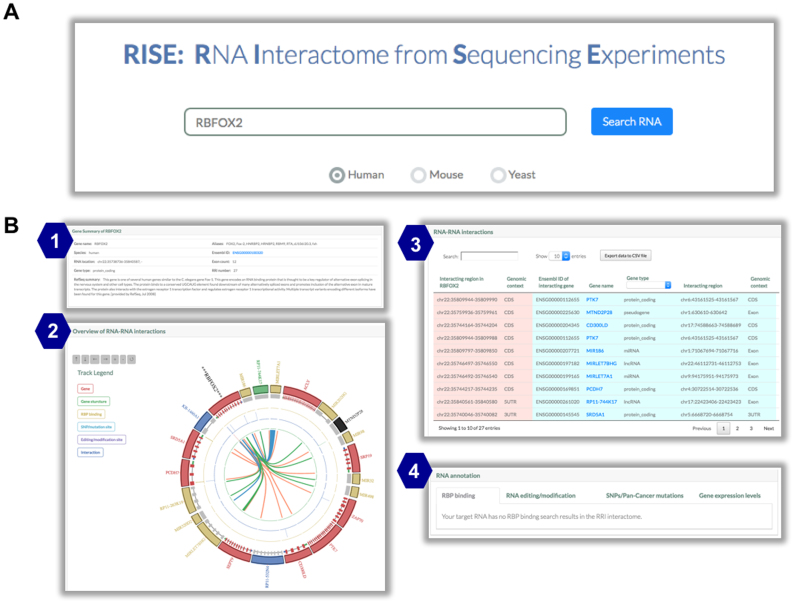
Web interface and example usage
We designed a user-friendly web interface to query the database. Users can input a gene name, e.g. RBFOX2, and choose a species (human, mouse or yeast) to search within (Figure 4A). RISE will return a table showing gene information and a Circos plot showing RRIs and associated annotations (Figure 4B). The Circos plot provides visualization of RRIs and multiple levels of annotations (Supplementary Figure S3), including (i) interacting gene name and type, (ii) gene structure (i.e. exon–intron structure of the longest transcript), (iii) RBP binding sites, (iv) SNPs and pan-cancer mutations, (v) RNA editing and modification sites and (vi) RNA interactions in the RISE database.
Figure 4.
Example of database usage: RBFOX2 in human as an example. (A) The search box of RISE. (B) The output of the search result showing: (1) gene information, (2) integrative view of RRIs and associated annotations in a Circos plot, (3) detailed information of RRIs, (4) associated molecular annotations.
Next, detailed information about each RRI involving RBFOX2 will be displayed in a table view (Figure 4B): the first two columns are about the interacting region on RBFOX2, the next five columns are about the interacting region on the RNA partner, and the last four columns are about the species, cell line, method/source and reference. Notably, RRIs collected from other databases do not have information of the interacting regions. Furthermore, users can navigate the molecular annotations of the RRIs (Figure 4B). Each time users can select one annotation module: (i) RBP binding, (ii) RNA editing/modification, (iii) SNPs/pan-cancer mutations or (iv) gene expression levels. In each module, RISE provides a table showing the information about molecular events located within the RRI regions. Users can download their search results by clicking on the ‘Export data to CSV file’ button. A more detailed explanation of the table view can be found on the Help web page.
DISCUSSION AND FUTURE DIRECTIONS
We present RISE, a comprehensive resource for RRIs identified through high-throughput sequencing technologies. Currently, RISE contains 328 811 RRIs mainly from human, mouse and yeast. The RISE database provides a convenient interface for RRI search and enables integrative navigation of RRIs with various molecular annotations. The major advantages of RISE over other similar databases (21–23) include (i) comprehensive curation of RRIs, (ii) a large dataset of RRIs among mRNAs and lncRNAs, (iii) details of the interacting sites and (iv) extensive annotations for each RRI.
Notably, when comparing different RRI datasets in RISE, we observed limited overlapping of RRIs among different experimental technologies even for the same cell line. As we discussed earlier, this could be partly explained by technology preferences and the issue of sequencing depth. It thus highlights the value of improving the technologies to be more sensitive and to achieve broader coverage on RRIs. Moreover, this limited overlap is also because the algorithms and parameters used to call interactions from sequencing data vary from technology to technology. The high degree of data heterogeneity poses difficulties in data reuse, integration and further discovery. It is thus highly desirable to develop a computational pipeline that standardizes experimental data processing and makes the results more comparable across technologies and studies.
We anticipate further development and improvement of the high-throughput sequencing-based technologies on RRI detection. It will generate more datasets and enable novel investigations into RRIs in various cellular and disease contexts. Some recent evidence has indicated linkage between dysregulation of RRIs and human diseases such as cancer (41) and genetic disorders (42). In addition, RRIs can be a new class of targets for drug discovery (43). As more data are generated in the future, we will maintain and keep updating RISE as a repository providing information and annotation on RRIs.
DATA AVAILABILITY
RISE is freely available at http://rise.zhanglab.net. The datasets in RISE can be downloaded and used in accordance with the GNU Public License and the license of their primary data sources.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Boqin Hu for technical advice on web construction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Natural Science Foundation of China [31671355]; National Thousand Young Talents Program of China (to Q.C.Z.). Funding for open access charge: National Natural Science Foundation of China National Thousand Young Talents Program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rinn J., Guttman M.. RNA Function. RNA and dynamic nuclear organization. Science. 2014; 345:1240–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferre F., Colantoni A., Helmer-Citterich M.. Revealing protein-lncRNA interaction. Brief. Bioinform. 2016; 17:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P.. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011; 146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guil S., Esteller M.. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem. Sci. 2015; 40:248–256. [DOI] [PubMed] [Google Scholar]
- 5. Consortium Encode Project. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L.. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011; 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P. et al. . Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009; 458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kung J.T., Colognori D., Lee J.T.. Long noncoding RNAs: past, present, and future. Genetics. 2013; 193:651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guttman M., Rinn J.L.. Modular regulatory principles of large non-coding RNAs. Nature. 2012; 482:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mercer T.R., Dinger M.E., Mattick J.S.. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009; 10:155–159. [DOI] [PubMed] [Google Scholar]
- 11. Cesana M., Daley G.Q.. Deciphering the rules of ceRNA networks. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7112–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Helwak A., Kudla G., Dudnakova T., Tollervey D.. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013; 153:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kudla G., Granneman S., Hahn D., Beggs J.D., Tollervey D.. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sugimoto Y., Vigilante A., Darbo E., Zirra A., Militti C., D’Ambrogio A., Luscombe N.M., Ule J.. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015; 519:491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K., Lee C.S., Flockhart R.J., Groff A.F., Chow J. et al. . Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013; 493:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engreitz J.M., Sirokman K., McDonel P., Shishkin A.A., Surka C., Russell P., Grossman S.R., Chow A.Y., Guttman M., Lander E.S.. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014; 159:188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu Z., Zhang Q.C., Lee B., Flynn R.A., Smith M.A., Robinson J.T., Davidovich C., Gooding A.R., Goodrich K.J., Mattick J.S. et al. . RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016; 165:1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aw J.G., Shen Y., Wilm A., Sun M., Lim X.N., Boon K.L., Tapsin S., Chan Y.S., Tan C.P., Sim A.Y. et al. . In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol. Cell. 2016; 62:603–617. [DOI] [PubMed] [Google Scholar]
- 19. Sharma E., Sterne-Weiler T., O’Hanlon D., Blencowe B.J.. Global mapping of human RNA-RNA interactions. Mol. Cell. 2016; 62:618–626. [DOI] [PubMed] [Google Scholar]
- 20. Nguyen T.C., Cao X., Yu P., Xiao S., Lu J., Biase F.H., Sridhar B., Huang N., Zhang K., Zhong S.. Mapping RNA-RNA interactome and RNA structure in vivo by MARIO. Nat. Commun. 2016; 7:12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hao Y., Wu W., Li H., Yuan J., Luo J., Zhao Y., Chen R.. NPInter v3.0: an upgraded database of noncoding RNA-associated interactions. Database (Oxford). 2016; 2016:baw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yi Y., Zhao Y., Li C., Zhang L., Huang H., Li Y., Liu L., Hou P., Cui T., Tan P. et al. . RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 2017; 45:D115–D118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Junge A., Refsgaard J.C., Garde C., Pan X., Santos A., Alkan F., Anthon C., von Mering C., Workman C.T., Jensen L.J. et al. . RAIN: RNA-protein association and interaction networks. Database (Oxford). 2017; 2017:baw167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai D., Meyer I.M.. A comprehensive comparison of general RNA-RNA interaction prediction methods. Nucleic Acids Res. 2016; 44:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao H., Sun Z., Wang J., Huang H., Kocher J.P., Wang L.. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014; 30:1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database (Oxford). 2011; 2011:bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hagberg A., Swart P., Swart D.. Exploring network structure, dynamics, and function using NetworkX. In Proceedings of the 7th Python in Science Conference (SciPy). 2008; 11–15. [Google Scholar]
- 30. Yang Y.C., Di C., Hu B., Zhou M., Liu Y., Song N., Li Y., Umetsu J., Lu Z.J.. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genomics. 2015; 16:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramaswami G., Li J.B.. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014; 42:D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kiran A., Baranov P.V.. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010; 26:1772–1776. [DOI] [PubMed] [Google Scholar]
- 33. Sun W.J., Li J.H., Liu S., Wu J., Zhou H., Qu L.H., Yang J.H.. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016; 44:D259–D265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang M.T., Asthana S., Gao S.P., Lee B.H., Chapman J.S., Kandoth C., Gao J., Socci N.D., Solit D.B., Olshen A.B. et al. . Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 2016; 34:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang Y., Yang Y.T., Yuan J., Lu Z.J., Li J.J.. Large-scale mapping of mammalian transcriptomes identifies conserved genes associated with different cell states. Nucleic Acids Res. 2017; 45:1657–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S. et al. . A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005; 122:957–968. [DOI] [PubMed] [Google Scholar]
- 39. Barabasi A.L., Oltvai Z.N.. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004; 5:101–113. [DOI] [PubMed] [Google Scholar]
- 40. Panni S., Prakash A., Bateman A., Orchard S.. Yeast non-coding RNA interaction network. RNA. 2017; 23:1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y., Xu X., Yu S., Jeong K.J., Zhou Z., Han L., Tsang Y.H., Li J., Chen H., Mangala L.S. et al. . Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017; 27:1112–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dusl M., Senderek J., Muller J.S., Vogel J.G., Pertl A., Stucka R., Lochmuller H., David R., Abicht A.. A 3′-UTR mutation creates a microRNA target site in the GFPT1 gene of patients with congenital myasthenic syndrome. Hum. Mol. Genet. 2015; 24:3418–3426. [DOI] [PubMed] [Google Scholar]
- 43. Matsui M., Corey D.R.. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017; 16:167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RISE is freely available at http://rise.zhanglab.net. The datasets in RISE can be downloaded and used in accordance with the GNU Public License and the license of their primary data sources.