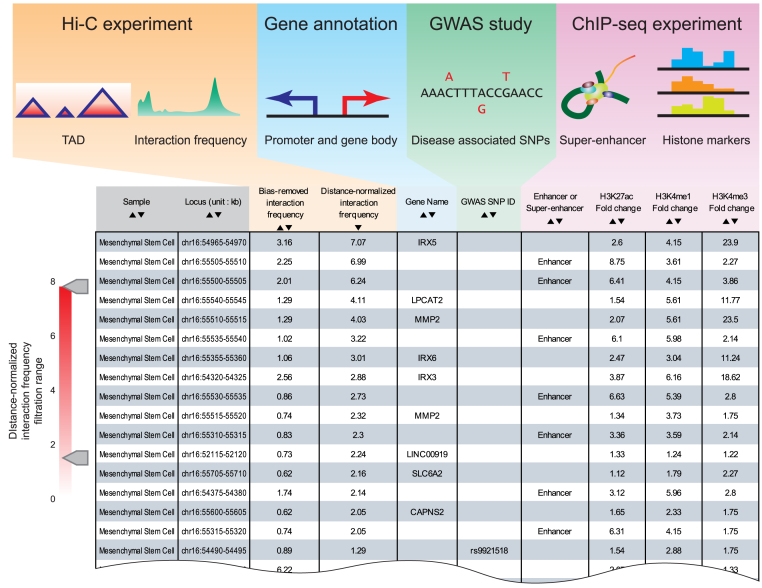
Figure 2.
Data sources and output result from interaction table module. 3DIV processed data from numerous sources including Hi-C experiments, gene annotations, GWAS studies and ChIP-seq experiments. From these processed data, 3DIV extracted chromatin interaction frequencies, annotation of promoters and gene bodies, coordinates of disease-associated SNPs, annotation of enhancer/super-enhancers and histone ChIP-seq signals. Interaction table provides a list of chromatin interaction partners for the queried locus with the aforementioned genomic/epigenomic annotations.