Abstract
Although successful in identifying new cataract-linked genes, the previous version of the database iSyTE (integrated Systems Tool for Eye gene discovery) was based on expression information on just three mouse lens stages and was functionally limited to visualization by only UCSC-Genome Browser tracks. To increase its efficacy, here we provide an enhanced iSyTE version 2.0 (URL: http://research.bioinformatics.udel.edu/iSyTE) based on well-curated, comprehensive genome-level lens expression data as a one-stop portal for the effective visualization and analysis of candidate genes in lens development and disease. iSyTE 2.0 includes all publicly available lens Affymetrix and Illumina microarray datasets representing a broad range of embryonic and postnatal stages from wild-type and specific gene-perturbation mouse mutants with eye defects. Further, we developed a new user-friendly web interface for direct access and cogent visualization of the curated expression data, which supports convenient searches and a range of downstream analyses. The utility of these new iSyTE 2.0 features is illustrated through examples of established genes associated with lens development and pathobiology, which serve as tutorials for its application by the end-user. iSyTE 2.0 will facilitate the prioritization of eye development and disease-linked candidate genes in studies involving transcriptomics or next-generation sequencing data, linkage analysis and GWAS approaches.
INTRODUCTION
While the identification of genes linked to eye development and its associated defects remains a challenge, the application of genome-level transcript profiling technologies to molecularly investigate specific eye tissues and cell-types promises to expedite this process (1,2). Nevertheless, high-throughput expression profiling brings yet new challenges, namely the analysis, compilation, access and visualization of the large amounts of data for prioritizing promising candidates for deeper analysis. We have previously addressed these challenges by developing a strategy termed ‘whole-embryo body (WB) in silico subtraction’ for prioritization of ocular disease genes and making this resource publicly accessible through a web-tool called iSyTE (integrated Systems Tool for Eye gene discovery) (1). Indeed, in silico subtraction has proven effective to prioritize genes in non-ocular tissues as well (3).
Development of the ocular lens is well characterized in various vertebrate model systems and discussed in detail elsewhere (4–6). Briefly, mouse lens development initiates when the optic vesicle interacts with the surface ectoderm and induces it to form the lens placode. The lens placode invaginates to form the lens pit that closes to form the lens vesicle. In subsequent stages, the posterior cells of the lens vesicle elongate and differentiate into primary fiber cells while the anterior cells form the lens epithelium. Throughout life, cells of the epithelium exit the cell cycle at the lens equator and differentiate into secondary fiber cells, which undergo terminal differentiation involving organelle degradation and expression of refractive proteins to form a transparent tissue. Over the past five years, the unbiased nature of the iSyTE prediction tool has led to new insights into fundamental regulatory mechanisms in ocular lens development and into the pathobiology of its associated disease, cataract. Significantly, iSyTE’s application has led to the identification of the post-transcriptional regulatory factor TDRD7 that is necessary for lens development in human, mouse and chicken (7)—a finding that opened up the investigation of RNA-binding protein mediated post-transcriptional control in lens development and cataract (8). Moreover, iSyTE has led to the identification and characterization of several other lens development and homeostasis genes including those encoding transcription factors (TFs) (Mafg, Mafk), cell adhesion proteins (Pvrl3) and selenoproteins (Sep15) that are associated with eye and/or lens defects (9–11), and has facilitated the analysis of several lens regulatory pathways (e.g.Crim1, Prox1, Sip1 (Zeb2), etc.) (12–15). In addition to impacting these eye development studies, iSyTE has expedited gene discovery in human cataract. For example, based on analysis of cataract-associated mapped intervals, iSyTE suggested that SIPA1L3 is linked to congenital cataracts in human (1). This prediction was subsequently validated by multiple reports that described SIPA1L3 mutations/deficiency to cause lens defects or cataract in human, mouse, frog and fish (16–18). Additionally, iSyTE prioritized several novel candidate genes for pediatric cataract (CYP51A1, GEMIN4, RIC1, TAF1A, TAPT1, WDR87) (19,20) and has provided supportive evidence in linking STX3 to congenital cataract (21). Furthermore, it has impacted the association of the causative genes in other human ocular disorders that concern the lens, including linking ADAMTS18 to microcornea and myopia and ASPH to Traboulsi syndrome (22,23).
While valuable, iSyTE’s previous version is limited to just three mouse lens developmental datasets and has limited capacity for data visualization, which is mainly restricted to lens-enrichment tracks in the UCSC (University of California Santa Cruz) Genome Browser (1). Presently, the eye research community has generated ∼140 microarray datasets on wild-type mouse lenses at various stages and on lens tissue from specific gene-perturbation mouse mutants that exhibit lens defects or cataract (2). The enormous potential of this data presently lies untapped in public databases such as GEO (Gene Expression Omnibus) and ArrayExpress (24,25). This is mainly because as currently deposited, the datasets are in the form of minimally processed (or unprocessed) files that need to be analyzed by the end-user to extract useful information. Further, there is no available resource that facilitates the analysis of new candidate gene(s) in the comprehensive context of all the existing wild-type or mutant lens expression data. Thus, to enhance iSyTE, as well as to make these under-utilized data effectively available to the research community, in this report we analyzed all the lens microarray gene expression datasets that have been generated using standard Affymetrix and Illumina platforms. We noted that the publicly available data was predominantly representative of mouse embryonic, early postnatal or adult stages, but largely lacking in mid-postnatal to two-month old adult stages. Therefore, we generated five new wild-type mouse lens microarray datasets to address this deficit. Moreover, to facilitate in silico subtraction analyses for determining lens-enriched genes from Illumina datasets, we generated new microarray data for mouse whole-embryo body (WB) on the Illumina WG-6 platform. Additionally, we developed a new iSyTE web interface that allows direct access and clear visualization of these thoroughly processed datasets while also facilitating a range of downstream analyses. We demonstrate the utility of the expanded iSyTE 2.0 database to identify and analyze genes and pathways associated with lens biology/pathobiology.
MATERIALS AND METHODS
Generation of new microarray datasets for iSyTE 2.0
Mice were housed at University of Delaware animal facility and animal experiments, approved by the Institutional Animal Care and Use Committee (IACUC), were performed following the guidelines in the Association of Research in Vision and Ophthalmology (ARVO) statement for the use of animals in ophthalmic and vision research. Wild-type ICR mice (Taconic) were used for generating new lens and WB microarray expression datasets (Supplementary Table S1). Four mouse lenses were used for each biological replicate and microarrays were performed on total RNA isolated using the RNeasy Mini Kit (Qiagen) at postnatal (P) stages P8, P12, P20, P42 and P52. Total RNA from mouse E10.5, E11.5 and E12.5 WB tissue (minus eyes) in equimolar ratios was used for generating the WB microarray dataset. Previously, we showed that WB datasets at different developmental stages are similarly proficient in the ‘subtraction’ process for identifying tissue-enriched genes (1). Therefore, to keep the comparisons uniform, we have used the above WB reference dataset for the subtraction of the lens datasets. Microarrays were performed on BeadChip MouseWG-6 v2.0 Expression arrays (Illumina) following described methods (26).
Microarray data analysis and implementation of iSyTE 2.0 database and web interface
Mouse lens microarray datasets at various stages were obtained from NCBI GEO database or generated new in this study (Supplementary Table S1) (1,7,9,12,27–33). These data on normal or specific gene-perturbation mouse lenses are on four different microarray platforms: Affymetrix 430 2.0; Affymetrix 430A 2.0; Illumina MouseWG-6 v1.0; Illumina MouseWG-6 v2.0. Data preprocessing and analyses were performed using the ‘R’ statistical environment with ‘affy’ and ‘lumi’ packages for the Affymetrix and Illumina microarray platforms, respectively. Principal component analysis (PCA) was performed for determining consensus between biological replicates. The outlying microarray samples were removed during data pre-processing as previously established (34). For Affymetrix, the datasets were imported, background corrected and normalized by ‘rma’ (Robust Multichip Average) algorithm using the ‘affy’ package. Batch effects were corrected using empirical Bayesian framework implemented as ‘ComBat’ function in Surrogate Variable Analysis package. Probes with a significant ‘mas5’ detection P-value (≤0.05) in less than three samples were filtered out. If multiple probe sets represented a single gene, the probe set with the highest median expression across all lens samples was used to represent its expression. Probes were annotated using latest annotations from ‘mouse4302.db’ package. For Illumina, datasets were imported, background corrected and normalized by the built-in ‘rankinvariant’ method using the ‘lumi’ package. Absent or low expressed probes were removed using the ‘detectionCall’ function of ‘lumi’ package such that only probes that were detected in at least two samples were retained for downstream analysis. If multiple probes represented one gene, the probe with the highest median expression across samples was used to represent it. Differential gene expression for mutants or lens-enrichment of genes by in silico WB-subtraction was estimated using ‘limma’ package using default parameters as described (1,26). Final output files, specific to microarray platform, were generated using ‘write.fit’ function implemented in ‘limma’. The iSyTE 2.0 database is implemented using MariaDB (version: 5.5.3) to support data retrieval and visualization. Processed data was uploaded to the database using custom scripts and the web-interface was built on LAMP (Linux, Apache, MySQL (now MariaDB), PHP) stack. The data is retrieved from MariaDB database through AJAX and presented to the user via DataTables jQuery plug-in. Color-coding of table is achieved using a custom heatmaps javascript, with styling by CSS (Cascading Style Sheet). The utility of the web interface is explained in detail in the ‘Results’ section.
Functional annotation clustering and gene regulatory network construction
The top 200 lens-enriched genes were selected from each developmental stage, as determined by lens versus WB comparison, this provided a non-redundant set of 528 lens-enriched genes from all lens stages on the Affymetrix 430 2.0 platform. These were used for further downstream analyses. Functional annotation clustering was performed using the DAVID bioinformatics resource (https://david.ncifcrf.gov/) with default settings. Expression-based clustering of genes was done using Self-organizing Tree Algorithm (SOTA) implemented in ‘ClValid’ package for ‘R’ statistical language with default parameters. The SOTA cluster genes (n = 528) exhibiting dynamic expression patterns across embryonic and postnatal lens development were investigated as follows. First, TF-binding motifs from the motif database MotifDb (http://bioconductor.org) for the TFs Pax6, Maf, Mafb, Mafg, Pitx3, Six6 and Sox1 were searched in the 2500 bp upstream TSS (transcription start site) of the 528 genes in the eleven SOTA clusters. The presence—and not enrichment—of TF-binding motifs in the upstream regions was analyzed. TF-binding motifs identified by matchPMW function in the Biostring package were used for this analysis. Next, we extracted edges for 528 genes from (i) STRING Db (http://string-db.org/) with default confidence score (>0.4) and (ii) from a co-expression based network that was generated using weighted correlation network analysis (WGCNA) package in R-statistical language (35); to generate this lens temporal gene regulatory network (GRN) we used all available lens perturbation data sets and followed the recommended protocol of data preparation and one-step network construction, and exported network to Cytoscape readable format using built-in function of WGCNA. In the expression-based network, the key parameter, soft thresholding power (β), for weighted network construction was optimized to maintain both the scale-free topology and sufficient node connectivity as recommended (35). Finally, an in-house python script was used for combining (i) the STRING interactions (edge score >0.4), (ii) the weighted correlation network edges (adjacency threshold >0.35) and (iii) the TF motif edges; the resulting integrated lens GRN was visualized using Cytoscape.
RESULTS
Analysis of mouse developmental and mutant lens microarrays for iSyTE 2.0
To construct iSyTE 2.0, we analyzed (and processed by WB-subtraction) all the available mouse lens microarray data generated on standard Affymetrix and Illumina platforms (Supplementary Table S1). Previously, we generated WB microarray data for the GeneChip™ Mouse Genome Affymetrix 430 2.0 platform to facilitate the in silico subtraction-based computation of lens-enrichment scores (1). However, there was no WB microarray dataset available that allows similar processing of lens datasets on the Illumina microchip. Therefore, we first generated a new WB microarray dataset on the Illumina MouseWG-6 v2.0 platform in this study. Armed with WB microarray data for both Affymetrix and Illumina platforms, we processed lens microarray data from wild-type mouse or those used as ‘controls’ in studies describing mutant-control comparisons for investigating gene expression dynamics in lens development from embryonic through postnatal stages. We noted that publicly available lens microarray datasets are primarily on embryonic/early postnatal stages and just three adult stages. To extend the representation of lens microarray data between postnatal through 2-month old adult stages, we generated five new wild-type mouse lens microarray datasets at stages P8, P12, P20, P42 and P52 (Supplementary Table S1, see ‘Materials and Methods’ section). PCA plots demonstrate that the lens samples form stage-related clusters i.e. embryonic or postnatal, and are distinct compared to the WB (Supplementary Figure S1A). This is also confirmed by correlation plots of the lens datasets that demonstrate their level of relatedness as a factor of developmental stage, displaying a varying consensus as would be expected from related developmental stages i.e. suggesting high-quality of our data (Supplementary Figure S1B).
Next, these datasets were processed by in silico subtraction using the WB datasets on the Affymetrix or Illumina platforms to determine lens-enriched genes. To test the efficacy of WB in silico subtraction in identifying genes that are functionally associated to lens biology, gene ontology (GO) associations were examined for the top 500 lens-enriched genes (lens versus WB comparison) for different stages. In the gene-set subjected to WB in silico subtraction GO terms directly related to lens biology such as ‘Lens-development in camera-type eye’ and ‘Lens fiber cell differentiation’ are enriched (Supplementary Figure S2A and B). In contrast, without WB in silico subtraction, the enriched GO terms were related to housekeeping function (Supplementary Figure S2A and B). Further, expression of genes linked to human cases of isolated (non-syndromic) cataract is significantly high in the lens compared to WB (Supplementary Figure S2C and D). This demonstrates the effectiveness of the WB in silico subtraction analysis to enrich lens-relevant genes from different microarray datasets regardless of platform or whether they are from embryonic, early postnatal or adult stages.
Next, we processed and performed differential gene expression analysis for all publicly available Illumina and Affymetrix lens microarray data on mouse mutants that exhibit lens defects or cataract (Supplementary Table S1). Specifically, data on the following mouse mutants with various gene perturbation conditions (germline or conditional knockout, dominant negative or ectopic overexpression) were considered: CBP:p300 double knockout at stages E9.5, E10.5; dominant negative mutant for Brg1 at stage E15.5; transgenic mutant with Cryaa promoter-driven Foxe3 overexpression in fiber cells at stage P2; Pax6 knockout at stages E9.5, E10.25, E10.5; E2f1:E2f2:E2f3 triple knockout at stages E17.5, P0; Notch2 knockout at stage E19.5; Hsf4 knockout at stage P0; Sparc knockout at stage P28 (isolated lens epithelium); Tdrd7 null at stages P4, P30; Klf4 knockout at stages E16.5, P56; Mafg−/−:Mafk+/− compound mutant at P60. The results of these analyses correlate well with previous findings on gene expression alterations in the various mutant conditions and are discussed in detail in later sections.
New iSyTE 2.0 web interface for lens gene expression visualization
To make this comprehensive resource accessible to the research community, we developed a user-friendly web interface (http://research.bioinformatics.udel.edu/iSyTE) that (i) provides effective visualization of the fully processed microarray data, and (ii) allows the end-user to perform various customized downstream analyses. We provide specific examples below to describe the utility of the iSyTE 2.0 web interface.
When the database is interrogated for the cataract-linked gene Tdrd7 using the ‘Mouse mm10’, ‘Normalized Expression’ in ‘All’ (stages) options, the output heat-map displays its elevated expression in the lens (Figure 1A–C, see legend for details), which correlates with initiation of fiber cell differentiation, as has been experimentally established (7). Further, iSyTE 2.0 offers several downstream analyses options such as the ‘Filter’ option to select genes based on normalized expression threshold or lens-enrichment cut-off for user selected lens stages (Figure 1D); the ‘Rank’ option to allow the ranking of genes in a gene list, on selected lens stages, based on expression or lens-enrichment (Figure 1D and E). In a comparison involving multiple stages the ‘rank’ option tool ranks genes based on the specific stage that shows the highest expression for a candidate gene (among multiple genes). Further, the ‘GO’ option (Figure 1D) allows GO-based mining of user-provided gene-set by connecting with the bioinformatics resource DAVID (Database for Annotation, Visualization and Integrated Discovery) (36).
Figure 1.
A new web-interface for iSyTE 2.0 database. (A) The fully analyzed Affymetrix Mouse 430 2.0 and Illumina MouseWG-6 v2.0 gene expression data from embryonic, postnatal lens development stages can be accessed under the tab ‘Lens Gene Expression’ on the iSyTE 2.0 web interface. For investigating the expression of candidate gene(s), under ‘Standard’, we developed a search portal called ‘Find expression data for’ where the user submits the gene query. (B) The query can be analyzed using specific parameters on the right as follows: (i) select the species (Mouse mm10 or Human hg19 assembly), (ii) select the type of comparison between the choices of (a) normalized expression, (b) lens enrichment and (c) mutant versus control, (iii) select the statistic appropriate for the comparison (e.g. for ‘lens enrichment’ the statistic choices offered are ‘fold change’, ‘t-stat’ or ‘P-value’) and (iv) selects the appropriate dataset (e.g. selecting ‘Developmental’ will show all normal lens expression data while selecting ‘Mutants’ will show all mutant lens expression data in fold-change; selection of specific mutants such as Brg1 is also an option), before submitting for analysis. (C) The output for the query gene(s) is provided as an expression heat-map for different mouse developmental stages. (D) iSyTE 2.0 offers various downstream analysis such as ‘Filter’, ‘Rank’ and ‘GO’ analysis via DAVID on selected genes. (E) In the following example, the ‘Rank’ feature is described. In an investigation of 14 candidate genes from an ∼6 Mb genomic region (chr16:77980000–83979999), iSyTE 2.0 lens expression or enrichment (fold change) correctly ranks the gene MAF as the top candidate, which is the causative gene linked to human congenital cataract. Additionally, tabs for other analysis/resources such as ‘Co-expression’, ‘Lens-enrichment in UCSC Brower’ and ‘Top Lens-enriched Genes’ are accessible under ‘Lens Gene Expression’.
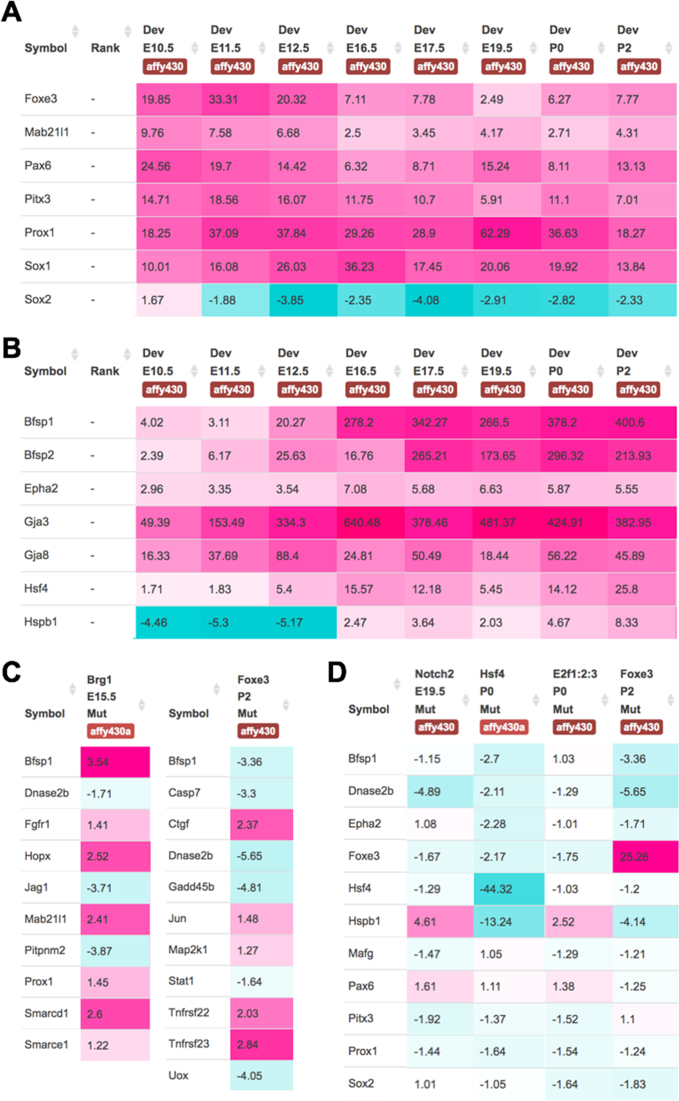
Several genes can be simultaneously interrogated through the new iSyTE 2.0 web interface. For example, examination of 10 crystallin genes shows their high expression and enrichment in the lens (Supplementary Figure S3A and B) and their developmentally relevant expression patterns. Namely, Cryaa and Cryab are highly lens-enriched from early stages of lens development (Supplementary Figure S3B) as described (37–39), while Crybb1 and Crygd exhibit progressively high expression and enrichment in later lens developmental stages as previously validated (40,41). Further, the iSyTE 2.0 also reflects the experimentally established patterns of regulatory genes in lens development. For example, lens-enriched expression of Foxe3 and Mab21l1 is progressively reduced in the developing whole lens tissue, whereas Prox1 expression becomes lens-enriched and correlates with fiber differentiation (Figure 2A). iSyTE 2.0 even captures a well-established expression switch in the TFs Sox2 and Sox1 (Figure 2A) (42–44) and shows the lens fiber differentiation-associated upregulation of the genes Bfsp1, Bfsp2, Epha2, Gja3, Gja8, Hsf4 and Hspb1 (Figure 2B), indicating the sensitivity of the database. It should be noted that absolute expression values of the same genes may be different between Affymetrix and Illumina platforms which reflects their inherent differences in probe sets (45). Users should avoid direct comparisons, especially in regard to absolute gene expression values, between the two platforms. Even within platforms, such comparisons should be limited to data generated on a specific type of microarray chip. Together, these findings serve to demonstrate that the integrated microarray analysis and the web-interface-enabled data visualization of the iSyTE 2.0 database allows investigation of gene expression patterns in normal lens development.
Figure 2.
iSyTE 2.0 allows effective visualization of normal and perturbed lens expression data. (A) Expression of lens regulators in different lens stages. At early lens development stages E10.5 Sox2 is enriched in the lens, but with the commencement of primary fiber cell differentiation at E11.5, it ceases to be lens-enriched while Sox1 becomes progressively lens-enriched. (B) Fiber differentiation associated genes that get sharply lens-enriched between E12.5 and E16.5 stages. (C) Differentially expressed genes in the Foxe3 overexpression and the Brg1 dominant-negative mutant lens. (D) Differential gene expression in Notch2, Hsf4, E2f1:E2f2:E2f3 and the Foxe3 mouse mutant lenses. Note: Hspb1 (Hsp27) is markedly downregulated in Hsf4 and Foxe3 mutants but upregulated in Notch2 and E2f1:E2f2:E2f3 mutants, among other differences in misregulated genes. It is also noteworthy that Foxe3 is 25-fold upregulated in the Foxe3 overexpression mutant while Hsf4 shows a 44-fold downregulation in the Hsf4 deletion mutant, indicating that iSyTE 2.0 meta-analysis has worked well.
Utility of iSyTE 2.0 for exploring cataract-associated differential gene expression
The new iSyTE 2.0 web-interface allows the user to investigate differential gene expression patterns in lenses from gene-perturbation mouse mutants with lens defects or cataract. For example, iSyTE 2.0-assisted investigation of the dominant-negative Brg1 mutant lenses shows that Bfsp1, Fgfr1, Hopx1, Mab21l1, Prox1, Smarcd1, Smarce1 are upregulated and Dnase2b, Jag1, Pitpnm2 are downregulated (Figure 2C), as previously described (27). Similarly, analysis of mutant lenses with Cryaa-promoter driven Foxe3 overexpression in fiber cells demonstrates that Bfsp1, Casp7, Dnase2b, Gadd45b, Stat1, Uox are downregulated and Ctgf, Jun, Map2k1, Tnfrsf22, Tnfrsf23 are upregulated (Figure 2C), in agreement with previous findings (28). Further, iSyTE 2.0 enables concurrent investigation of multiple mutant datasets. For example analysis of Notch2 deletion, Hsf4 deletion, E2f1:E2f2:E2f3 triple deletion and Foxe3 overexpression mouse mutants reveals previously unappreciated differences in mis-expressed genes (Figure 2D). This example shows that although fiber cell defects may appear phenotypically similar in different mutants, iSyTE 2.0 analysis can reveal distinct molecular signatures that may influence the manifestation of these defects. Further, an option called ‘Fold change with P-value’ under ‘Select statistic shown’ allows visualization of fold-change data with statistical significance for Mutant versus Control comparisons. Thus, iSyTE 2.0 offers an integrated environment for systematic investigation of multiple candidate genes, simultaneously, in several different gene perturbation mouse mutants with lens defects or cataract.
Application of iSyTE 2.0 for prioritization of cataract-linked genes
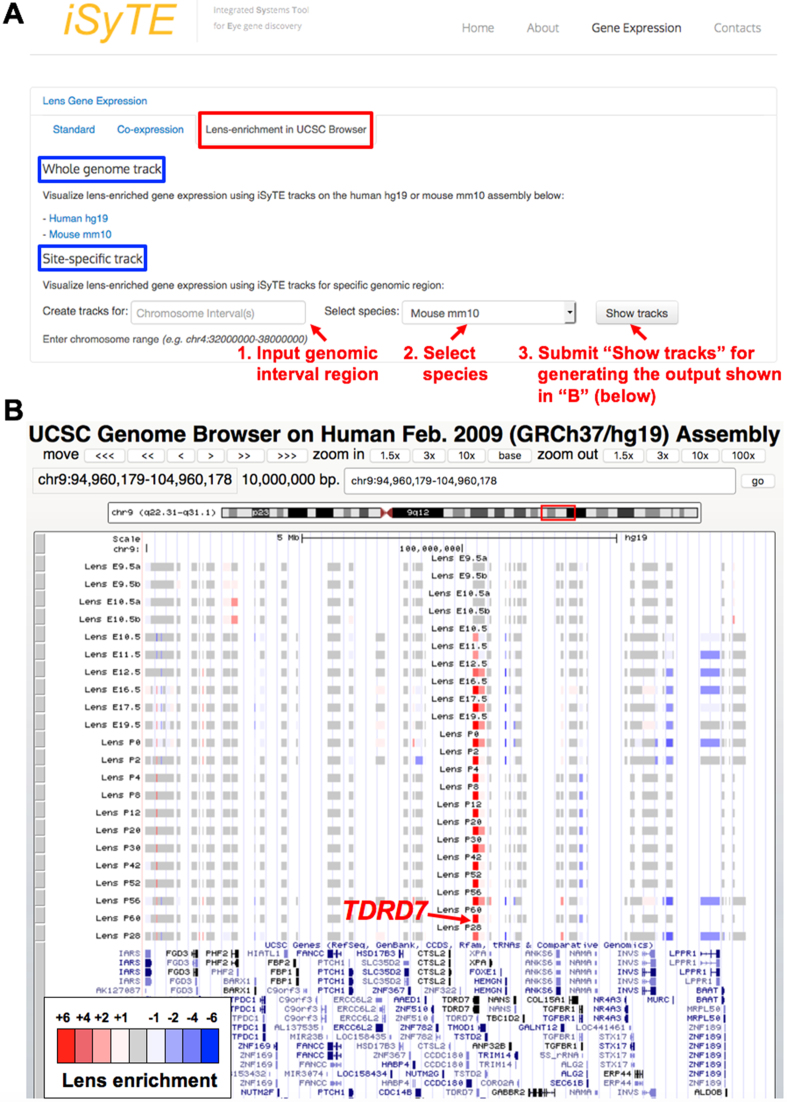
To effectively apply lens gene expression profiles for prioritization of candidate genes related to lens biology and cataract, several new features are now introduced in iSyTE 2.0. First, lens-enrichment for individual genes can now be viewed in the context of the mouse genome GRCm38/mm10 assembly or the human genome GRCh37/hg19 assembly using the UCSC Genome Browser. On the iSyTE 2.0 website, ‘Lens-enrichment on UCSC Browser’ tab is now provided for direct access to the UCSC Genome Browser displaying twenty-two iSyTE custom tracks that show lens-enrichment heat-maps at different developmental stages (Figure 3A and B). Additionally, as described, a new ‘Rank’ feature is provided under the ‘Standard’ tab to order a gene-list (e.g. from patient high-throughput sequencing data or from a mapped linkage-interval) by first selecting the lens stage and then ordering it on lens enrichment t-statistic or fold change (Figure 1E).
Figure 3.
iSyTE 2.0 offers custom tracks to visualize lens-enrichment in UCSC Genome Browser. (A) Application of iSyTE 2.0 tracks for visualizing mouse gene expression data on the mouse genome GRCm38/mm10 assembly or the human genome (GRCh37/hg19) assembly in the UCSC Genome browser. This feature enables the user to visualize lens gene expression information in the broad context of various other informative resources (such as evolutionary conservation, ENCODE data, etc.) available at the UCSC Genome browser. On the main iSyTE 2.0 website, the tab ‘Lens-enrichment on UCSC Browser’ provides access to the UCSC Genome Browser via ‘Whole genome track’ or ‘Site-specific track’ (for direct access to specific region of interest), which display 22 iSyTE custom tracks. (B) iSyTE lens-enrichment tracks provide heat-maps (intense red corresponds to high lens-enrichment) corresponding to various mouse lens developmental stages for candidate genes, which can be viewed in the mouse genome GRCm38/mm10 assembly or the human genome GRCh37/hg19 assembly. As an example, lens-enrichment for TDRD7 in a 10 Mb interval on human chr9 is shown.
The inclusion of multiple lens developmental stages and the gene-perturbation lens data in the new iSyTE 2.0 increases its utility in prioritization of cataract-linked genes. This is illustrated by the investigation of three cataract-linked genes CHMP4B, FYCO1, GCNT2 that were not effectively identified by the previous iSyTE version (1). We had predicted that this was due to the limited embryonic stages (n = 3) on the previous iSyTE, and that these genes are likely enriched at later stages of lens development (1). In agreement, the updated iSyTE 2.0 shows that these genes are indeed lens-enriched at later stages (Supplementary Figure S4A). Further, these genes exhibit altered expression in lenses of various gene-perturbation mouse mutants with cataract (Supplementary Figure S4B). Finally, the iSyTE 2.0 ‘Rank’ feature shows an improvement of ranks for all three candidates among the genes present in their mapped intervals (Supplementary Figure S4C–E). Collectively, these findings highlight the efficacy of the updated iSyTE 2.0 features for prioritization of cataract-linked candidate genes.
Utility of iSyTE 2.0 in identification of lens-signature genes and regulators
iSyTE 2.0 presents an opportunity to recognize gene expression data representative of embryonic and/or postnatal lens stages. To investigate this further, we used platform-specific datasets to infer embryonic and postnatal lens molecular signatures. This analysis identified 49 genes broadly segregated into four groups displaying distinct temporal expression (Supplementary Figure S5). While this lens signature gene-set includes some expected genes (e.g. crystallins), it also highlights new candidates for future studies, namely Aldoc, Dhx32, Fabp5, Gprc5b, Grifin, Gstm1, Hmgn3, Mboat1, Mocs2, Npl, Ogn, Pgam2, Tmem40 and Zbtb8b.
iSyTE 2.0 for gaining insights into temporal dynamics of lens gene expression
iSyTE 2.0 can be applied to identify high-priority candidates based on co-expression dynamics across embryonic and postnatal lens stages. The web interface offers a ‘Co-expression’ query feature that allows user to identify the top candidate genes with an expression pattern similar to a gene of interest, for example Cryga (Supplementary Figure S6). Co-expression analysis using iSyTE 2.0 is effective because the database contains comprehensive lens expression data across multiple stages. The utility of iSyTE 2.0 in this regard is demonstrated by the following detailed analyses that reveals biologically relevant information.
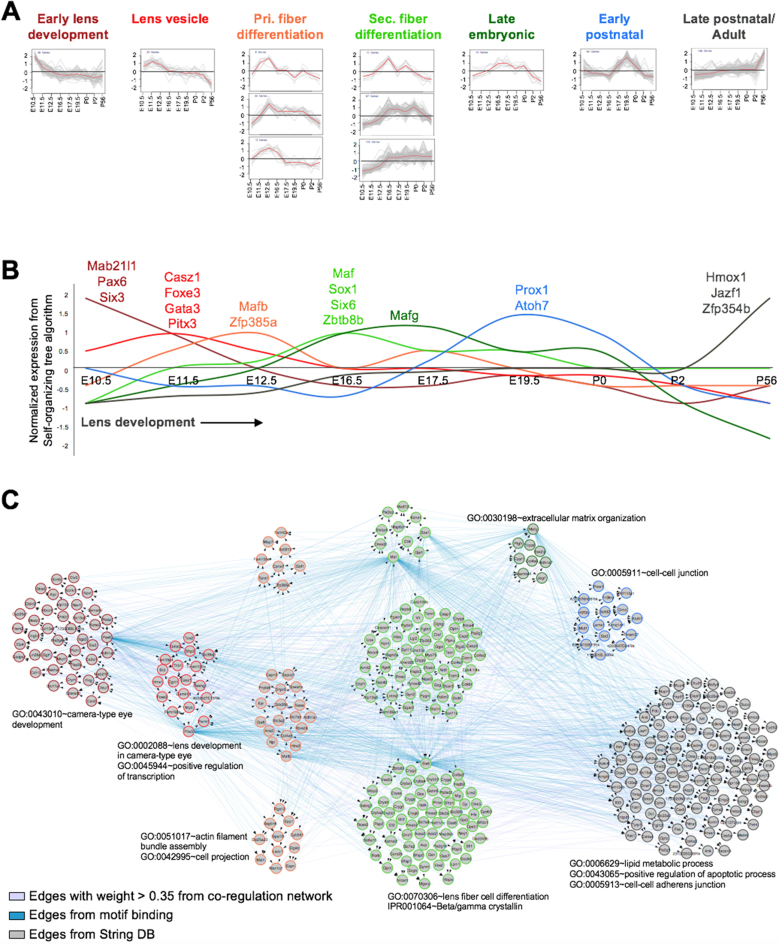
Expression-based clustering analysis (SOTA, see ‘Materials and Methods’ section) on the top 200 lens-enriched genes in all the Affymetrix lens datasets identifies eleven gene expression clusters (Figure 4A and Supplementary Table S2). The genes considered for this analysis can be viewed and downloaded using the tab ‘Top Lens-enriched Genes’ under ‘Lens Gene Expression’. These clusters exhibit distinct patterns for genes co-regulated in the lens, with peak expression at stages that correlate to ‘Early lens development’ (Cluster 3, 56 genes), ‘Lens vesicle’ (Cluster 4, 20 genes), ‘Primary fiber differentiation’ (Cluster 5, 22 genes; cluster 10, 8 genes and cluster 11, 12 genes), ‘Secondary fiber differentiation’ (Cluster 6, 103 genes; cluster 7, 87 genes and cluster 8, 10 genes), ‘Late embryonic’ (Cluster 9, 10 genes), ‘Early postnatal’ (Cluster 2, 62 genes) and ‘Late postnatal/Adult’ (Cluster 1, 138 genes) (Figure 4A). This analysis identifies known as well as potential new TFs in the lens. For example, genes with peak expression at the early lens development stage include the established genes Pax6, Mab21l1 and Six3 (Figure 4B), while the cluster corresponding to the lens vesicle stage contains Foxe3 and Pitx3. The primary and secondary fiber differentiation stage clusters contain Mafb, Maf and Sox1, while Mafg and Prox1 are identified in the gene clusters that exhibit peak expression at the late-embryonic and early postnatal stages. Moreover, this analysis identifies Casz1, Gata3, Hmox1, Jazf1, Six6, Zbtb8b, Zfp354b and Zfp385a as new TF candidates in the lens (Figure 4B). Further investigation reveals the interconnectivity between the candidate genes in these eleven clusters (Figure 4C; Supplementary Figures S7 and 8). Together, these findings show how iSyTE 2.0 can assist in the identification of regulatory factors to provide new insights into lens biology.
Figure 4.
iSyTE 2.0 provides insights into lens expression dynamics. (A) Expression based clustering (SOTA clustering) of the top 200 lens-enriched genes from mouse stages E10.5, E11.5, E12.5, E16.5, E19.5, P0, P2 and P56 identified eleven clusters, which are classified based on their dynamic expression patterns and peak expression stage. Clusters are plotted such that X-axis represents normalized expression in SOTA, while Y-axis shows lens development stages. The red line in each plot depicts mean expression pattern of genes in a specific cluster across different stages. (B) Combined expression dynamics of TF-genes across all eleven lens development stage clusters (‘Early lens development’ (dark red), ‘Lens vesicle’ (red), ‘Pri. fiber differentiation’ (orange), ‘Sec. fiber differentiation’ (light green), ‘Late embryonic’ (dark green), ‘Early postnatal’ (blue) and ‘Late postnatal/Adult’ (black). TFs that exhibit peak expression in each cluster are indicated. (C) A combined network of eleven cluster genes (n = 528) derived from WGCNA correlation network (purple edges), transcription factor binding motif analysis (blue edges) and String DB (gray edges) across the different developmental stages. Node border color represent specific clusters.
CONCLUSION AND FUTURE WORK
This report describes the meta-analysis of all available lens microarray datasets on two commonly used platforms and their integration into an updated iSyTE 2.0 database with a new user-friendly web interface. Further it also describes how iSyTE 2.0 can be applied to analyze new candidate genes or gene datasets to gain insights into lens development and homeostasis. iSyTE 2.0 is constructed to enable visualization and allow comparative analyses of gene expression data across various mouse developmental lens stages, while also showing how specific gene perturbations cause alterations in these expression profiles. The new iSyTE 2.0 interface provides information on lens-expression (absolute values), lens-enrichment metric (fold-change or t-statistic), seamless integration with GO resource (DAVID), functionality to identify co-expressed genes, integrated visualization through lens-enrichment tracks on the UCSC Browser and differential gene expression profiles for all publicly available gene-perturbation mouse mutants with cataract. Importantly, although the microarray data are generated from different mouse backgrounds and laboratory conditions, the overall lens gene expression profiles from the iSyTE 2.0 meta-analysis correlate well with established lens gene expression patterns that are previously validated by in situ hybridization and immunofluorescence analysis.
We expect that the new iSyTE 2.0 resource will have a far-reaching impact on identification of lens development and disease associated genes. In particular, because iSyTE 2.0 allows the end-user to simultaneously analyze any new candidate gene data in the context of these comprehensive lens expression data, it would greatly impact prioritization of candidates from patient next-gen sequencing analysis, mapped intervals, and GWAS studies. Indeed, future use of iSyTE 2.0 with other resources such as DECIPHER (Database of Chromosomal Imbalance and Phenotype in Humans using Ensemble Resources; http://decipher.sanger.ac.uk/) (46)—that provide information on human copy number variants linked to various phenotypes including cataract, as well as the cataract-gene database Cat-Map (47) can expedite the identification of new cataract-linked genes. Future iSyTE updates will include RNA-sequencing and protein data and expression information on other ocular tissues. Finally, iSyTE 2.0 is expected to find broader utility among developmental biologists and clinical geneticists outside of the eye field by assisting in the identification of genes that are linked to multiple tissue disorders in syndromic cases that involve the eye.
CITING iSyTE 2.0
The following citation format is suggested when referring to data obtained from iSyTE 2.0: these data were retrieved from iSyTE 2.0 (integrated Systems Tool for Eye gene discovery, URL: http://research.bioinformatics.udel.edu/iSyTE). To reference the database, please cite this article.
DATA AVAILABILITY
The newly generated mouse lens microarray data reported here have been deposited with the Gene Expression Ontology Database at NCBI under accession number GSE100136.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Cathy Wu for her support in hosting the iSyTE web-resource at the Center for Bioinformatics and Computational Biology at the University of Delaware. The authors dedicate this work to the memory of Dr David C. Beebe, Ph.D., FARVO, Janet and Bernard Becker Professor of Ophthalmology and Visual Sciences, Washington University, St Louis, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01EY021505 to S.A.L.]; The Pew Charitable Trusts Biomedical Sciences Scholarship (to S.A.L.). Funding for open access charge: National Institutes of Health [R01EY021505 to S.A.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lachke S.A., Ho J.W.K., Kryukov G.V., O’Connell D.J., Aboukhalil A., Bulyk M.L., Park P.J., Maas R.L.. iSyTE: integrated Systems Tool for Eye gene discovery. Invest. Ophthalmol. Vis. Sci. 2012; 53:1617–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anand D., Lachke S.A.. Systems biology of lens development: a paradigm for disease gene discovery in the eye. Exp. Eye Res. 2017; 156:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu H., Busch T., Eliason S., Anand D., Bullard S., Gowans L.J.J., Nidey N., Petrin A., Augustine-Akpan E.-A., Saadi I. et al. . Exome sequencing provides additional evidence for the involvement of ARHGAP29 in Mendelian orofacial clefting and extends the phenotypic spectrum to isolated cleft palate. Birth Defects Res. 2017; 109:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Donner A.L., Lachke S.A., Maas R.L.. Lens induction in vertebrates: variations on a conserved theme of signaling events. Semin. Cell Dev. Biol. 2006; 17:676–685. [DOI] [PubMed] [Google Scholar]
- 5. Lachke S.A., Maas R.L.. Building the developmental oculome: systems biology in vertebrate eye development and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010; 2:305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cvekl A., Ashery-Padan R.. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014; 141:4432–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lachke S.A., Alkuraya F.S., Kneeland S.C., Ohn T., Aboukhalil A., Howell G.R., Saadi I., Cavallesco R., Yue Y., Tsai A.C.-H. et al. . Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011; 331:1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dash S., Siddam A.D., Barnum C.E., Janga S.C., Lachke S.A.. RNA-binding proteins in eye development and disease: implication of conserved RNA granule components. Wiley Interdiscip. Rev. RNA. 2016; 7:527–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Agrawal S.A., Anand D., Siddam A.D., Kakrana A., Dash S., Scheiblin D.A., Dang C.A., Terrell A.M., Waters S.M., Singh A. et al. . Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum. Genet. 2015; 134:717–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lachke S.A., Higgins A.W., Inagaki M., Saadi I., Xi Q., Long M., Quade B.J., Talkowski M.E., Gusella J.F., Fujimoto A. et al. . The cell adhesion gene PVRL3 is associated with congenital ocular defects. Hum. Genet. 2012; 131:235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kasaikina M.V., Fomenko D.E., Labunskyy V.M., Lachke S.A., Qiu W., Moncaster J.A., Zhang J., Wojnarowicz M.W. Jr, Natarajan S.K., Malinouski M. et al. . Roles of the 15-kDa selenoprotein (Sep15) in redox homeostasis and cataract development revealed by the analysis of Sep 15 knockout mice. J. Biol. Chem. 2011; 286:33203–33212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolf L., Harrison W., Huang J., Xie Q., Xiao N., Sun J., Kong L., Lachke S.A., Kuracha M.R., Govindarajan V. et al. . Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res. 2013; 41:10199–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manthey A.L., Lachke S.A., FitzGerald P.G., Mason R.W., Scheiblin D.A., McDonald J.H., Duncan M.K.. Loss of Sip1 leads to migration defects and retention of ectodermal markers during lens development. Mech. Dev. 2014; 131:86–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Audette D.S., Anand D., So T., Rubenstein T.B., Lachke S.A., Lovicu F.J., Duncan M.K.. Prox1 and fibroblast growth factor receptors form a novel regulatory loop controlling lens fiber differentiation and gene expression. Development. 2016; 143:318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y., Fan J., Ho J.W.K., Hu T., Kneeland S.C., Fan X., Xi Q., Sellarole M.A., de Vries W.N., Lu W. et al. . Crim1 regulates integrin signaling in murine lens development. Development. 2016; 143:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evers C., Paramasivam N., Hinderhofer K., Fischer C., Granzow M., Schmidt-Bacher A., Eils R., Steinbeisser H., Schlesner M., Moog U.. SIPA1L3 identified by linkage analysis and whole-exome sequencing as a novel gene for autosomal recessive congenital cataract. Eur. J. Hum. Genet. 2015; 23:1627–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greenlees R., Mihelec M., Yousoof S., Speidel D., Wu S.K., Rinkwitz S., Prokudin I., Perveen R., Cheng A., Ma A. et al. . Mutations in SIPA1L3 cause eye defects through disruption of cell polarity and cytoskeleton organization. Hum. Mol. Genet. 2015; 24:5789–5804. [DOI] [PubMed] [Google Scholar]
- 18. Rothe M., Kanwal N., Dietmann P., Seigfried F.A., Hempel A., Schütz D., Reim D., Engels R., Linnemann A., Schmeisser M.J. et al. . An Epha4/Sipa1l3/Wnt pathway regulates eye development and lens maturation. Development. 2017; 144:321–333. [DOI] [PubMed] [Google Scholar]
- 19. Aldahmesh M.A., Khan A.O., Mohamed J.Y., Hijazi H., Al-Owain M., Alswaid A., Alkuraya F.S.. Genomic analysis of pediatric cataract in Saudi Arabia reveals novel candidate disease genes. Genet. Med. 2012; 14:955–962. [DOI] [PubMed] [Google Scholar]
- 20. Patel N., Anand D., Monies D., Maddirevula S., Khan A.O., Algoufi T., Alowain M., Faqeih E., Alshammari M., Qudair A. et al. . Novel phenotypes and loci identified through clinical genomics approaches to pediatric cataract. Hum. Genet. 2017; 136:205–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chograni M., Alkuraya F.S., Ourteni I., Maazoul F., Lariani I., Chaabouni H.B.. Autosomal recessive congenital cataract, intellectual disability phenotype linked to STX3 in a consanguineous Tunisian family. Clin. Genet. 2015; 88:283–287. [DOI] [PubMed] [Google Scholar]
- 22. Aldahmesh M.A., Alshammari M.J., Khan A.O., Mohamed J.Y., Alhabib F.A., Alkuraya F.S.. The syndrome of microcornea, myopic chorioretinal atrophy, and telecanthus (MMCAT) is caused by mutations in ADAMTS18. Hum. Mutat. 2013; 34:1195–1199. [DOI] [PubMed] [Google Scholar]
- 23. Patel N., Khan A.O., Mansour A., Mohamed J.Y., Al-Assiri A., Haddad R., Jia X., Xiong Y., Mégarbané A., Traboulsi E.I. et al. . Mutations in ASPH cause facial dysmorphism, lens dislocation, anterior-segment abnormalities, and spontaneous filtering blebs, or Traboulsi syndrome. Am. J. Hum. Genet. 2014; 94:755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. . NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y.A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T. et al. . ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 2015; 43:D1113–D1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anand D., Agrawal S., Siddam A., Motohashi H., Yamamoto M., Lachke S.A.. An integrative approach to analyze microarray datasets for prioritization of genes relevant to lens biology and disease. Genome Data. 2015; 5:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He S., Pirity M.K., Wang W.-L., Wolf L., Chauhan B.K., Cveklova K., Tamm E.R., Ashery-Padan R., Metzger D., Nakai A. et al. . Chromatin remodeling enzyme Brg1 is required for mouse lens fiber cell terminal differentiation and its denucleation. Epigenet. Chromatin. 2010; 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Landgren H., Blixt A., Carlsson P.. Persistent FoxE3 expression blocks cytoskeletal remodeling and organelle degradation during lens fiber differentiation. Invest. Ophthalmol. Vis. Sci. 2008; 49:4269–4277. [DOI] [PubMed] [Google Scholar]
- 29. Gupta D., Harvey S.A.K., Kenchegowda D., Swamynathan S., Swamynathan S.K.. Regulation of mouse lens maturation and gene expression by Krüppel-like factor 4. Exp. Eye Res. 2013; 116:205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang J., Rajagopal R., Liu Y., Dattilo L.K., Shaham O., Ashery-Padan R., Beebe D.C.. The mechanism of lens placode formation: a case of matrix-mediated morphogenesis. Dev. Biol. 2011; 355:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wenzel P.L., Chong J.-L., Sáenz-Robles M.T., Ferrey A., Hagan J.P., Gomez Y.M., Rajmohan R., Sharma N., Chen H.-Z., Pipas J.M. et al. . Cell proliferation in the absence of E2F1-3. Dev. Biol. 2011; 351:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saravanamuthu S.S., Le T.T., Gao C.Y., Cojocaru R.I., Pandiyan P., Liu C., Zhang J., Zelenka P.S., Brown N.L.. Conditional ablation of the Notch2 receptor in the ocular lens. Dev. Biol. 2012; 362:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Greiling T.M.S., Stone B., Clark J.I.. Absence of SPARC leads to impaired lens circulation. Exp. Eye Res. 2009; 89:416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oldham M.C., Konopka G., Iwamoto K., Langfelder P., Kato T., Horvath S., Geschwind D.H.. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008; 11:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langfelder P., Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008; 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang D.W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 37. Zwaan J. The appearance of alpha-crystallin in relation to cell cycle phase in the embryonic mouse lens. Dev. Biol. 1983; 96:173–181. [DOI] [PubMed] [Google Scholar]
- 38. Robinson M.L., Overbeek P.A.. Differential expression of alpha A- and alpha B-crystallin during murine ocular development. Invest. Ophthalmol. Vis. Sci. 1996; 37:2276–2284. [PubMed] [Google Scholar]
- 39. Haynes J.I., Duncan M.K., Piatigorsky J.. Spatial and temporal activity of the alpha B-crystallin/small heat shock protein gene promoter in transgenic mice. Dev. Dyn. 1996; 207:75–88. [DOI] [PubMed] [Google Scholar]
- 40. Duncan M.K., Li X., Ogino H., Yasuda K., Piatigorsky J.. Developmental regulation of the chicken beta B1-crystallin promoter in transgenic mice. Mech. Dev. 1996; 57:79–89. [DOI] [PubMed] [Google Scholar]
- 41. Tréton J.A., Jacquemin E., Courtois Y., Jeanny J.C.. Differential localization by in situ hybridization of specific crystallin transcripts during mouse lens development. Differentiation. 1991; 47:143–147. [DOI] [PubMed] [Google Scholar]
- 42. Nishiguchi S., Wood H., Kondoh H., Lovell-Badge R., Episkopou V.. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 1998; 12:776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Donner A.L., Episkopou V., Maas R.L.. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev. Biol. 2007; 303:784–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Donner A.L., Ko F., Episkopou V., Maas R.L.. Pax6 is misexpressed in Sox1 null lens fiber cells. Gene Expr. Patterns. 2007; 7:606–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barnes M., Freudenberg J., Thompson S., Aronow B., Pavlidis P.. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005; 33:5914–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P.. DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 2009; 84:524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shiels A., Bennett T.M., Hejtmancik J.F.. Cat-Map: putting cataract on the map. Mol. Vis. 2010; 16:2007–2015. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The newly generated mouse lens microarray data reported here have been deposited with the Gene Expression Ontology Database at NCBI under accession number GSE100136.