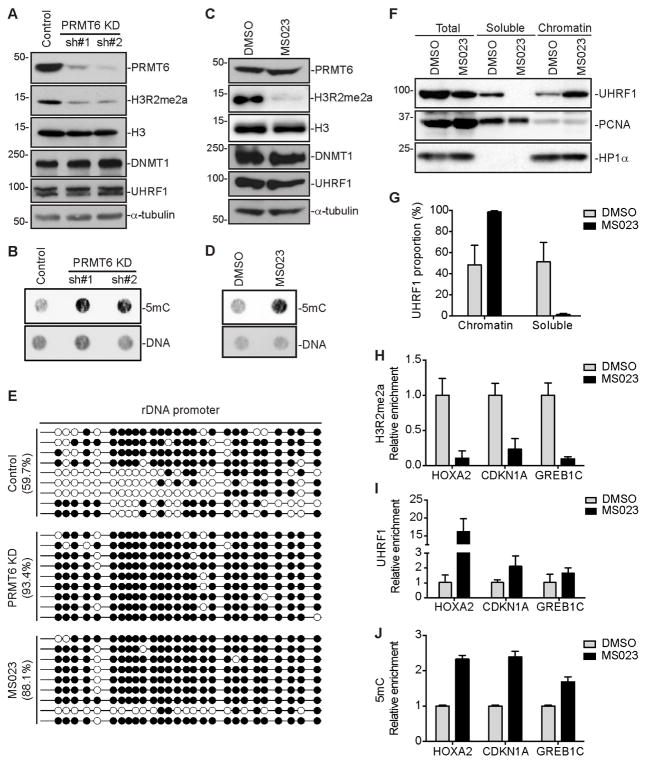
Figure 4. PRMT6 depletion or inhibition restores DNA methylation in MCF7 cells.
(A and C) Western blot analysis of MCF7 cells stably transfected with PRMT6 shRNAs (A) or treated with MS023 (C).
(B and D) 5mC dot blot analysis of samples in (A) and (C), respectively. DNA loading was verified by SYTOX Green staining.
(E) Bisulfite sequencing analysis of a 45S rDNA promoter region containing 27 CpG sites. Open circles, unmethylated CpGs; Filled circles, methylated CpGs.
(F) Nuclear fractionation of MCF7 cells treated with MS023 for UHRF1 chromatin association. PCNA and HP1α were used as controls for soluble and chromatin-associated proteins, respectively.
(G) Quantification of data in (F) by densitometry using Image J. Shown are percentages of soluble and chromatin-associated UHRF1 in each sample (mean + SD from three independent experiments).
(H–J) ChIP or MeDIP assays showing relative enrichment of H3R2me2a (H) and UHRF1 (I) or relative DNA methylation levels (J) at HOXA2, CDKN1A and GREB1C promoter regions (mean + SD from three independent experiments).
MS023 treatment was performed at 10 μM for 4 days for all experiments.
See also Figure S4.