Abstract
Background
Dystrophin, an essential protein functional in the maintenance of muscle structural integrity is known to be responsible for muscle deterioration during white spot syndrome virus (WSSV) infection among prawn species. Previous studies have shown the upregulation of dystrophin protein in Macrobrachium rosenbergii (the giant freshwater prawn) upon white spot syndrome virus (WSSV) infection. The literature has also suggested the important role of calcium ion alterations in causing such muscle diseases. Thus, the interest of this study lies within the linkage between dystrophin functioning, intracellular calcium and white spot syndrome virus (WSSV) infection condition.
Methods
In this study, the dystrophin gene from M. rosenbergii (MrDys) was first characterised followed by the characterization of dystrophin gene from a closely related shrimp species, Penaeus monodon (PmDys). Dystrophin sequences from different phyla were then used for evolutionary comparison through BLAST analysis, conserved domain analysis and phylogenetic analysis. The changes in mRNA expression levels of dystrophin and the alteration of intracellular calcium concentrations in WSSV infected muscle cells were then studied.
Results
A 1246 base pair long dystrophin sequence was identified in the giant freshwater prawn, Macrobrachium rosenbergii (MrDys) followed by 1082 base pair long dystrophin sequence in P. monodon (PmDys). Four conserved domains were identified from the thirteen dystrophin sequences compared which were classified into 5 different phyla. From the phylogenetic analysis, aside from PmDys, the characterised MrDys was shown to be most similar to the invertebrate phylum of Nematoda. In addition, an initial down-regulation of dystrophin gene expression followed by eventual up-regulation, together with an increase in intracellular calcium concentration [Ca2+]i were shown upon WSSV experimental infection.
Discussion
Both the functionality of the dystrophin protein and the intracellular calcium concentration were affected by WSSV infection which resulted in progressive muscle degeneration. An increased understanding of the role of dystrophin-calcium in MrDys and the interactions between these two components is necessary to prevent or reduce occurrences of muscle degeneration caused by WSSV infection, thereby reducing economic losses in the prawn farming industry from such disease.
Keywords: Infectious disease, Immunology
1. Introduction
Dystrophin is a large and complex gene responsible for maintaining the structural integrity of muscle fibres (Muntoni et al., 2003). Furthermore, dystrophin, and the dystrophin-associated proteins dystroglycan and sarcoglycan, work together in muscular contraction within the cytoskeleton. However, mutations in dystrophin or dystrophin-associated genes can lead to destabilization of the muscle structure and subsequently to muscular dystrophy, a progressive muscle wasting disorder (Rybakova et al., 2000). Additionally, intracellular calcium ion [Ca2+]i is involved in the regulation and modulation of muscle contractions and other muscle related activities such as protein metabolism, differentiation and growth. Studies have further shown that alterations in [Ca2+]i concentrations are often associated with muscle diseases, lending support to the idea that [Ca2+]i are important muscle signalling molecules (Goonasekera et al., 2014; Mallouk et al., 2000). Elevation of the total calcium content of dystrophy affected muscles has been observed in cases of muscle degeneration disorders, membrane damage and disturbances to calcium homeostasis (Miyake and McNeil, 2003; Ruegg and Gillis, 1999).
Another interesting point is revealed in an earlier report by Rao et al. (2016) concerning the up-regulation of the muscle-related gene, dystrophin in WSSV infected Macrochium rosenbergii. White spot syndrome virus (WSSV) infection, which leads to white spot disease (WSD), is one of the most common diseases to affect the aquaculture industry over the past two decades. This virus has been proven to be pathogenic to a wide range of temperate crustacean decapods, including the giant freshwater prawn or Macrobrachium rosenbergii (Corteel et al., 2012; Pradeep et al., 2012; Hameed et al., 2003; Kiran et al., 2002). M. rosenbergii infected with WSSV displays a range of symptoms, including reduction in feed uptake, loose cuticles, discoloration of muscles and appendages as well as blatant muscular lethargy (Bateman et al., 2012). WSSV virus infection presumably affects dystrophin gene expression and [Ca2+]i concentrations, which then causes muscular lethargy.
Taking into account of Rao et al.’s (2016) findings and the highly conserved nature of the dystrophin gene throughout the animal kingdom, M. rosenbergii is a good candidate for the study of the changes of dystrophin upon WSSV infection. In line with this, the aim of our study was to examine the changes in mRNA expression of dystrophin upon WSSV infection in M. rosenbergii. We also sought to quantify changes in intracellular calcium ion [Ca2+]i in order to study correlations between dystrophin and calcium ion in maintaining the structural integrity of muscle fibres.
2. Materials and methods
2.1. Identification, amplification and characterization of Macrobrachium rosenbergii dystrophin (MrDys) sequence
Macrobrachium rosenbergii dystrophin (MrDys) short sequences were obtained from the NCBI Sequence Read Archive under the accession numbers SRR1424572 and SRR1424575. The sequences encoding the dystrophin gene were confirmed with BLAST homology against the NCBI database (http://blast.ncbi.nlm.nih.gov). Gene-specific primers (DysF 5′-GGTCTCAGGGGACAAAATGA-3′) and (DysR 5′-TGGGGTGAGTGATCTTGTGA-3′) were designed to target the MrDys. The verification of MrDys was conducted through PCR amplifications with the designed primers and subsequent sequencing, using healthy prawn DNA. The reaction was performed in 15 μl reactions containing DNA template (50 ng/μl), 1 × Mastermix (Applied Biosystems, USA), DysF (0.30 μM) and DysR (0.30 μM) respectively. The thermal profile used was 95 °C for 10 min and 45 cycles of 95 °C for 15 s, 57.8 °C for 30 s, and 72 °C for 75 s. The amplified product was sequenced through Sanger sequencing on an ABI 3730XL DNA Analyzer (Applied Biosystems, USA) and its corresponding amino acid sequence was deduced through computational translation using the ExPASy protein translation tool (http://expasy.org/tools/dna.html).
2.2. Identification, amplification and characterization of Penaeus monodon dystrophin (PmDys) sequence
Based on the Macrobrachium rosenbergii dystrophin (MrDys) sequence obtained previously through PCR amplification and Sanger Sequencing, primers sets (PMD1F 5′-AGATCAACACGCCCACA-3′), (PMD1R 5′-GCTACGAACTGATGGTTCAATG-3′), (PMD2F 5′-TGGGAGAGGGCTACAACTA-3′) and (PMD2R 5′-CACCGCTGACACATATCAAAG-3′) were designed to target the dystrophin gene of P. monodon (PmDys). The PmDys was then verified through PCR amplifications followed by sequencing. A total of 50 μl mixture was used per PCR reaction including DNA template (50 ng/μl), EasyTaq Buffer (with MgCl2), EasyTaq Polymerase, dNTP (2.5 μM), PMD1F (10 μM), PMD1R (10 μM), PMD2F (10 μM) and PMD2R (10 μM) respectively. The thermal profile used was 95 °C for 5 min, 35 cycles of 95 °C for 45 s, 51.6 °C for 45 s, 72 °C for 30 s and a final extension of 72 °C for 5 min. The amplified product was sequenced through Sanger sequencing on an ABI 3730XL DNA Analyzer (Applied Biosystems, USA) and its corresponding amino acid sequence was deduced through computational translation using the ExPASy protein translation tool (http://expasy.org/tools/dna.html).
2.3. Conserved domain and phylogenetic analysis
Nucleotide sequences of the gene from eleven different organisms selected from 5 different phyla (Chordata, Nematoda, Echinodermata, Mollusca and Arthrophoda) were retrieved from the NCBI database. The sequences obtained were translated into protein sequences. These protein sequences, as well as MrDys and PmDys, were analysed against the NCBI conserved domain database (Marchler-Bauer et al., 2011) to identify their conserved regions, and a graphic comparison of protein domains was produced. A phylogenetic analysis of the thirteen dystrophin protein sequences (including MrDys and PmDys) was conducted through the Neighbour-joining Maximum Likelihood method, using the MEGA 7 software (Tamura et al., 2013). A phylogenetic tree was constructed, with the number of substitutions per site used to measure branch lengths.
2.4. Experimental animals
Specific-pathogen-free freshwater prawns, Macrobrachium rosenbergii (5 g to 8 g), were obtained from a farm in Kedah, Peninsular Malaysia, and delivered to the laboratory at the University of Malaya in Kuala Lumpur. DNA from the muscle tissue of the prawns was extracted using the DNAeasy extraction kit (Qiagen, USA). The extraction was conducted on individual three prawns at each time point studied, with three replicates for each prawn. The concentration and purity of the extracted DNA were checked using a NanoDrop 2000 Spectrometer (Thermo Scientific, USA) at 260 nm and 280 nm prior to use in PCR. The DNA of the prawns was screened with the virus-specific PCR primers, VP28-140Fw (5′-AGGTGTGGAACAACACATCAAG-3′) and VP28-140Rv (5′-TGCCAACTTCATCCTCATCA-3′) (Mendoza-Cano and Sánchez-Paz, 2013). The amplification was performed in 15 μl reactions containing the DNA template (50 ng/μl), 1 × Mastermix (Applied Biosystems, USA) and 0.30 μM of each of the above two primers. The thermal profile used was 95 °C for 10 min, and 45 cycles of 95 °C for 15 s and of 60 °C for 1 min. Randomly selected prawns were dissected and the prawn muscle tissue samples were verified through PCR to have no WSSV infection. The prawns were then acclimatised for 7 days in 300 L flat-bottomed glass tanks (27° C). A total of 10 prawns were placed in each tank throughout the study. The prawns were fed once a day with commercially available prawn feed (Red Bee Aquarium Shrimp Feed, China).
2.5. Quantification of WSSV copy numbers
A WSSV viral standard with a known copy number (6.62 × 106 copies/μl) was obtained from BioSatria (Sabah, Malaysia). A 10-fold serial dilution ranging from 6.62 × 1010 copies/μl to 6.62 × 106 copies/μl of the viral DNA filtrate was carried out prior to quantitative PCR analysis. The quantitative PCR reactions were carried out using SYBR Green supermix (Applied Biosystems, USA) on an Applied Biosystems 7500 Real-Time PCR System. The amplifications were performed in 15 μl reactions containing viral DNA template (6.62 × 1010 copies/μl to 6.62 × 106 copies/μl), 1 × SYBRgreen Mastermix (Applied Biosystems, USA), VP28-140Fw (0.30 μM) and VP28-140Rv (0.30 μM). The qPCR was initiated with a single step of 95 °C for 10 min, followed by a 45-cycle thermal profile of 95 °C for 10 min and 60 °C for 1 min. The cycle threshold (Ct) values of the reaction were calculated by the inbuilt ABI 7500 SDS software. In accordance with the Mendoza-Cano methodology (Mendoza-Cano and Sánchez-Paz, 2013), a standard curve was constructed using the Ct values obtained from the qPCR of the serial diluted viral standard. This standard curve was later employed to determine the virus copy number at each stage of infection following immune challenge.
2.6. Virus preparation
The white spot syndrome virus (WSSV) used in this study was extracted from muscle tissues of infected Penaeus monodon samples. The infected tissue was homogenized in 0.1 g/ml TN buffer (20 mM Tris–HCl, 400 mM NaCl; pH 7.4) and centrifuged at 2000 × g for 10 min. The isolated virus was diluted with NaCl (1%, w/v) in a ratio of 1:5 (v/v), and filtered using a 0.45 μm syringe filter. The filtrate was stored at −80 °C prior to injection in the prawn samples. The copy number of the viral filtrate was determined through the research of Mendoza-Cano and Sánchez-Paz obtained from the qPCR (as described earlier) and compared with a plot of the standard curve. The copy number of the virus load was diluted to 2.7 × 106 copies/ml.
2.7. Immune challenge of prawns with WSSV
A total of 5 ml of viral filtrate (2.7 × 106 copies/ml) was intramuscularly injected into the fourth abdomen of each prawn, using a sterile syringe. Prior to this, five prawn samples were separated into individual tanks as a control group. Phosphate saline buffer (5 ml) was injected into each control sample. Three individual prawns were randomly collected at the following intervals: 0 hours, 3 hours, 6 hours, 12 hours, 24 hours, 36 hours and 48 hours post infection. The samples were anaesthetized and dissected in sterile conditions before being snap-frozen in liquid nitrogen and subjected to total RNA extraction.
2.8. Total RNA isolation and cDNA conversion
Total RNA was isolated from muscle tissue (50 mg). The samples’ muscle tissues were snap-frozen in liquid nitrogen prior to extraction using TRIzol® Reagent (Life Technologies, Carlsbad, USA). The extracted RNA was treated with RNAse-free DNAse (5 Prime GmbH, Hamburg, Germany) to eliminate DNA contamination. The total extracted total RNA (4 ul) was then reverse transcribed by GoScript reverse transcriptase (Promega, WI, USA) and Oligo(dT)15 Primer (Promega, WI, USA). The concentration and purity of the cDNA were checked before being used to quantify gene expression, using a NanoDrop 2000 Spectrometer (Thermo Scientific, USA) at 260 nm and 280 nm.
2.9. Quantification of MrDys expression
The expression of MrDys at different post-infection hours was determined by a quantitative PCR using SYBR Green supermix (Applied Biosystems, USA) on an Applied Biosystems 7500 Real-Time PCR System. The qPCR was carried out using the converted cDNA as a template, and with gene-specific primers (MrDysZZ fw 5′-TAGCTGTTTTGCATCGTGTTG-3′ and MyDysZZ rv 5′-TGGGGTGAGTGATCTTGTGA-3′). The amplifications were performed in 15 μl reactions containing cDNA template (50 ng/μl), 1 × SYBRgreen Mastermix (Applied Biosystems, USA), MrDysZZ fw (0.30 μM) and MyDysZZ rv (0.30 μM). The thermal profile consisted of an initial step at 50 °C for 2 min, followed by 95 °C for 10 min and 45 cycles of 95 °C for 15 s and 60 °C for 1 min. Cycle threshold (Ct) values were calculated by the inbuilt ABI 7500 SDS software. The specificity of the qPCR amplification was verified through a melt curve analysis by generating a dissociation curve. An internal control gene, elongation factor 1-alpha ELF-1 (Dhar et al., 2009), was quantified using the above reaction mixture with two different primers: ELF-1 specific primers Fw (5′-CGCCGAACTGCTGACCAAGA-3′; 0.30 μM) and ELF-1 specific primers Rv (5′-CCGGCTTCCAGTTCCTTACC-3′; 0.30 μM). The relative gene expression of the MrDys gene compared to the internal control gene was calculated in accordance with the comparative CT method (2–ΔΔCT) (Livak and Schmittgen, 2001). For each time interval in qPCR, 3 biological replicates (3 prawns per time interval) were used together with 3 technical replicates for each biological replicate for experimental validation purpose.
2.10. Quantification of intracellular Ca2+ concentration, [Ca2+]i
A fluorescent calcium chelator colorimetric quantification kit, or Calcium Assay Kit (Metallogenics, CHB, JAP), was used to quantify the amount of intracellular calcium ions present in the muscle tissues of the prawn samples. The muscle tissue (50 mg) was first submerged in a 3% Trichloroacetic Acid (TCA) solution. It was then crushed, using a sterile microcentrifuge tube homogenizer, and vortexed vigorously prior to incubation at 4 °C for 30 min. The mixture was centrifuged for 15 min at 10000 × g. The supernatant was collected for further analysis. The absorbance of the supernatant was calculated by a microplate reader (TECAN M200 PRO, USA) at 570 nm and 700 nm. The [Ca2+]i was measured using a standard calibration curve for calcium solution (10 mg/dL) (Jin et al., 2007). The calcium concentration (mg/dL) was calculated using the following equation:
2.11. Transmission Electron Microscope (TEM) analysis of M. rosenbergii muscle tissues
For the comparison of healthy and WSSV infected M. rosenbergii muscle tissues, tissue samples of infected M. rosenbergii at 24 hours and 48 hours post WSSV infection were prepared using a gradient dehydration method. The dissected muscle tissues were trimmed into 3 mm3 cubes and fixed in 4% glutaraldehyde (buffered in 0.1 M cacodylate buffer) for 4 hours at 4 °C. The cubes were then gently washed in 0.1 M cacodylate buffer for twice.
The washed tissue cubes were immersed in 1% osmium tetroxide (buffered in 0.1 M cacodylate buffer) for 2 hours at 4 °C for fixation. After the immersion, washing with double distilled water was needed to remove any traces of the osmium tetroxide fixing reagent prior to gradual dehydration. Gradual dehydration of the sample tissues were carried out by soaking them in a series of ethanol solution with increasing concentrations, including 35%, 50%, 70% and 95% through mixing with ultrapure distilled water. The dehydration was carried out in ascending order of ethanol solution concentrations, for 10 minutes at each concentration. Finally, the tissue cubes were dehydrated 3 times in pure ethanol (100%) for 15 minutes each.
The dehydrated tissue cubes were placed in propylene oxide solution for 15 minutes, and this step was repeated twice. Next, the tissues were infiltrated with a mixture of propylene oxide and resin solution at the ratio of 1:1 for 1 hour. This step was repeated with propylene oxide and resin infiltration at a ratio of 1:3 for 2 hours, followed by overnight soaking of tissues in pure resin. The fixated samples were then gently placed in specimen vials and rotated on a rotator at room temperature. Subsequently, the tissue cubes were embedded in capsules filled with resin and allowed to polymerize overnight at 60 °C in a heated chamber.
The polymerized blocks were then trimmed and cut into semi-thin sectioning using a glass knife and stained with methylene blue. Next, ultra-thin sections with a thickness of approximately 70 nm were cut using a diamond knife in a RMC PT-PC Powertome Ultramicrotome. The ultra-thin sections produced were stored in a vacuum container until transmission electron microscope (TEM) viewing. Before viewing, the prepared ultra-thin sections of the tissue were placed gently and precisely on a copper grid. The samples were then viewed using a HT7700 TEM (Hitachi, Japan) at 100 kV accelerating voltage in High Contrast Mode. Appropriate zooming and focusing options were selected for each viewing, and images of infected and healthy M. rosenbergii muscle fibers were captured using a charge-coupled device camera.
2.12. Statistical analysis
All of the quantifications were conducted in three biological replicates, each with three technical replicates. All the numerical data obtained were analysed using a one-way ANOVA and Dunnett’s multiple comparison tests at a significance level of P < 0.05, using GraphPad Prism 6 software (California, USA). The data is represented as a mean ± standard error means (S.E.M.) for each set of readings.
3. Results
3.1. Identification and characterization of Macrobrachium rosenbergii dystrophin (MrDys)
A 1246 base pair (bp) dystrophin sequence was obtained through the amplification of M. rosenbergii cDNA (Supplementary Fig. 1a) and submitted to the GenBank database under accession number KU198993. The nucleotide sequence of MrDys was translated into an amino acid sequence using the Expasy Translate Tool (Supplementary Fig. 1b). The calculated molecular weight of the MrDys protein was 30 kDa, with an isoelectric point (p.I) of 8.57. A homology analysis against the NCBI database revealed a close similarity of MrDys with dystrophin isoforms from various other organisms, including P. monodon (86%), Acromyrmex echinatior (71%), Halyomorpha halys (71%), Ceratosolen solmsi marchali (70%), Apis mellifera (69%), Bemisia tabaci (68%) and Fopius arisanus (67%).
3.2. Identification and characterization of Penaeus monodon dystrophin (PmDys)
A 1082 base pair (bp) dystrophin sequence was obtained through the amplification of P. monodon cDNA (Supplementary Fig. 2a). The nucleotide sequence of PmDys was translated into an amino acid sequence using the Expasy Translate Tool (Supplementary Fig. 2b). The calculated molecular weight of the PmDys protein was 28 kDa, with an isoelectric point (p.I) of 8.35. During a homology analysis against the NCBI database, a high similarity was shown between the PmDys and dystrophin isoforms from other organisms, such as M. rosenbergii (86%), Fopius arisanus (73%), Athalia rosea (71%), Acromyrmex echinatior (71%), Ceratosolen solmsi marchali (71%), Neodiprion lecontei (68%) and Bemisia tabaci (67%) which was similar to the homology analysis results of MrDys.
3.3. Conserved domain and phylogeny
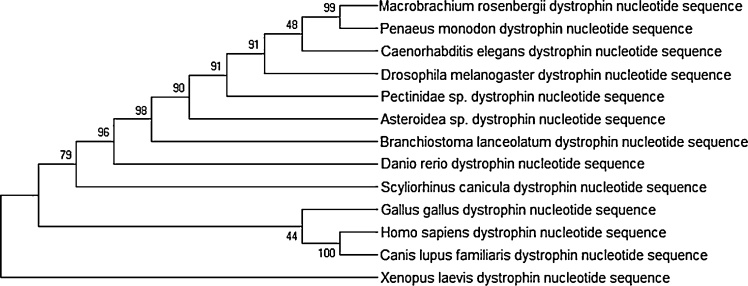
The translated MrDys (Supplementary Fig. 3a) and PmDys (Supplementary Fig. 3b) sequences showed four conserved domains: Spectrin, WW, EF Hand DMD-Like and ZZ superfamily domain. A study of homologous dystrophin sequences from eleven other different organisms – Gallus gallus, Homo sapiens, Canis lupus familiaris, Xenopus laevis, Scyliorhinus caniculua, Danio relio, Branchiostoma lancealatum, Asteriodea sp., Pectinidae sp., Drosophila melanogaster, and Caenorhabditis elegans – showed the presence of similar conserved domains to those in MrDys and PmDys (Supplementary Figure 4). The conserved domain analysis revealed that vertebrates display more repeats in the Spectrin domain than invertebrates which adds additional complexity to the vertebrate dystrophin genes (Supplementary Figure 4).
A phylogenetic tree, based on the maximum likelihood, was built to further investigate the relationship between MrDys and the dystrophin sequences of the twelve other organisms (Fig. 1). This showed MrDys clustered together with dystrophin homologues from P. monodon, the nematode C. elegans and the arthropod D. melanogaster. This suggests the close evolutionary relationship between the dystrophin genes of invertebrates.
Fig. 1.
Phylogenetic tree of dystrophin nucleotide sequences from 13 different organisms, including MrDys and PmDys, obtained by the Maximum Likelihood method using MEGA 7. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. MrDys was closely related to the dystrophin of Penaeus monodon, Caenorhabditis elegans and Drosophila melanogaster.
3.4. WSSV infection of M. rosenbergii
Infection symptoms were not observable in the early stages (0–12 hours) after the injection of WSSV into the abdomen of healthy M. rosenbergii. At 12 hours post infection, the prawns gradually began to grow lethargic and exhibit a loss of appetite. Other clinical symptoms caused by WSSV (discoloration of muscle and loose cuticles) were observed at 24 hours post infection. The infection symptoms continued to grow until 48 hours post infection. No mortality was observed, although at 48 hours post infection, the M. rosenbergii appeared to be highly moribund with debilitated swimming abilities observed with naked eyes.
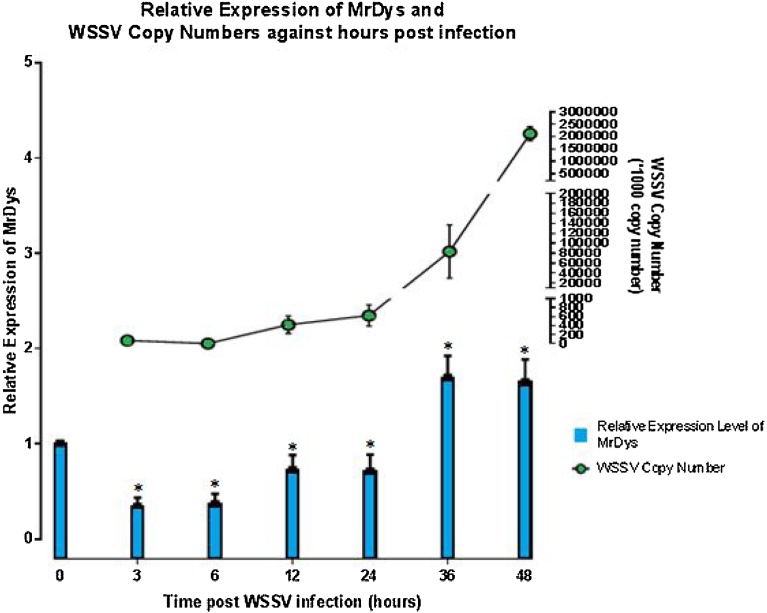
A quantification of the WSSV copy numbers in the infected M. rosenbergii muscle tissues similarly showed no significant changes until 24 hours post infection (Fig. 2). The copy numbers of WSSV at 3, 6 and 24 hours post-infection were 5.47 × 104, 2.95 × 103 and 4.99 × 105 copies/μl, respectively. However, the presence of WSSV increased drastically to 5.4 × 107 copies at 36 hours post infection, and continued to increase exponentially to 2.1 × 109 copies/μl at 48 hours post infection (Fig. 2).
Fig. 2.
Analysis of dystrophin-like gene expression in M. rosenbergii (MrDys) in response to WSSV infection by qPCR.
The qPCR on both MrDys and WSSV were carried out at intervals of 2, 6, 12, 24, 36 and 48 hours post infection, with a significant difference of (P < 0.05). The mRNA levels of MrDys were analysed and standardized relative to the reference gene (ELF1) expression. The viral load in each sample at the corresponding time point was quantified through absolute qPCR, and the results are shown in the line graph. Each bar and point represents the mean value from three replicates, while the error bars represent the standard error. In sum, MrDys and the virus copy number both began to increase sharply at 36 hours post infection.
3.5. Expression of MrDys in muscle tissue
The expression level of MrDys in muscle tissues in the infected sample group fell 0.33 fold and 0.36 fold at 3 and 6 hours post-infection respectively, compared to the levels in the uninfected control group (Fig. 2). The expression level of MrDys then gradually increased from 12 hours post infection onwards, rising to 1.6 fold higher than in the control sample group at 36 hours post infection. It then remained stable until 48 hours post infection.
3.6. Intracellular calcium concentration, [Ca2+]i, in muscle tissue
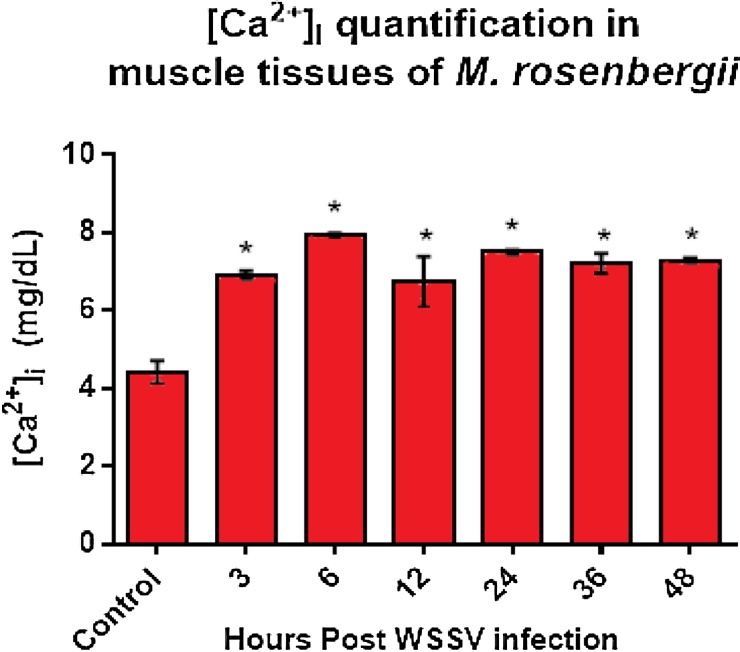
The amount of intracellular calcium [Ca2+]i in the muscle tissue of the WSSV infected M. rosenbergii showed a significant increase at 3 hours post WSSV infection, from 4.5 mg/dL in the control samples to 6.9 mg/dL (Fig. 3). A further increase in [Ca2+]i was observed until a peak of 7.9 mg/dL at 6 hours post infection. The Ca2+ concentration then fell to 6.75 mg/dL at 12 hours post infection, before rising again slightly and then fluctuating, with levels of 7.5 mg/dL, 7.22 mg/dL and 7.29 mg/dL at 24 hours, 36 hours and 48 hours post-injection, respectively.
Fig. 3.
Quantification of intracellular calcium, [Ca2+]i in M. rosenbergii muscle tissue.
The results were collected at 3, 6, 12, 24, 36 and 48 hours post WSSV infection. The concentrations shown were obtained from three technical replicates at each time point, calibrated with a 10 mg/dL calcium solution. The error bar shown is the standard error between mean values obtained at each time point. Asterisks indicate significant differences (P < 0.05) in [Ca2+]i between the infected and control tissue samples. Overall, a 1.6 fold increase in intracellular calcium was observed in the infected samples compared to the control samples.
3.7. Transmission Electron Microscope (TEM) imaging of muscle tissues
M. rosenbergii muscle sample tissues from the following time points of the WSSV immune challenge were used in TEM imaging of muscle morphology: 0 hours (control), 24 hours and 48 hours post infection. These muscle tissue samples were dissected and processed as elaborated in the methodology for each time point. The images taken at each time point were shown in Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9 . Qualitative comparison of the images obtained was conducted for a clear comparison of muscle morphology of the sample tissues under different conditions, focusing on muscle fibers and mitochondria. The comparisons showed that tissues obtained from the control samples were the least intact, while samples at 48 hours post WSSV infection were affected the most.
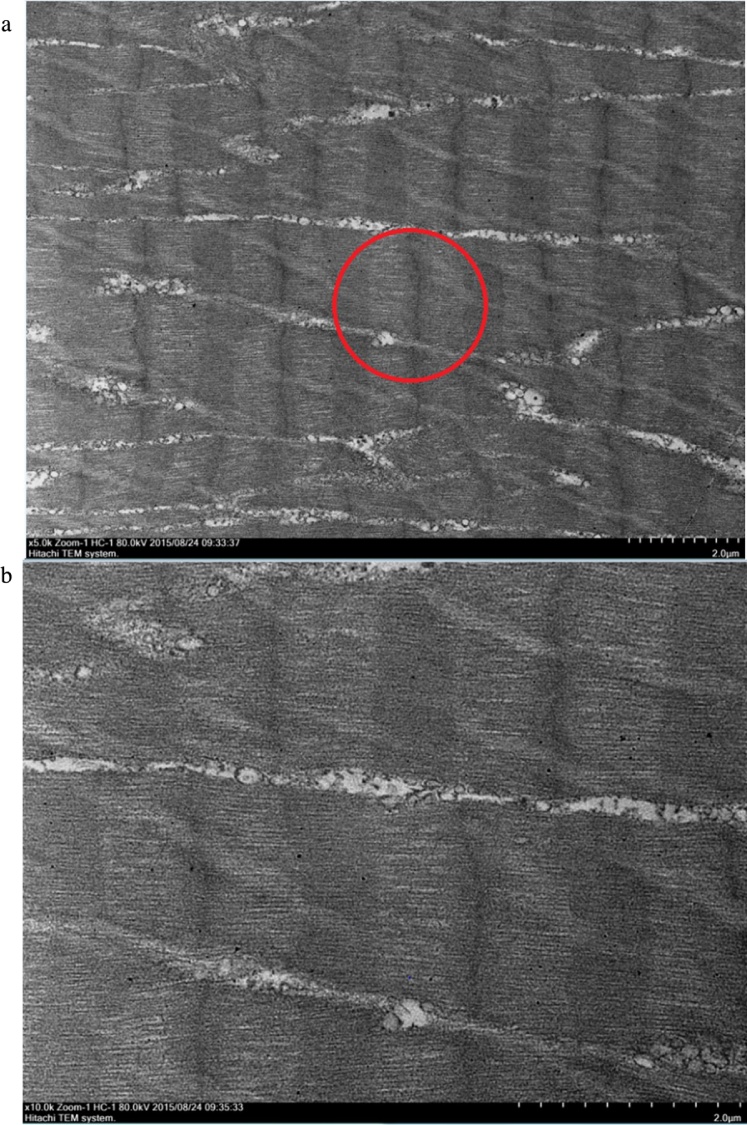
Fig. 4.
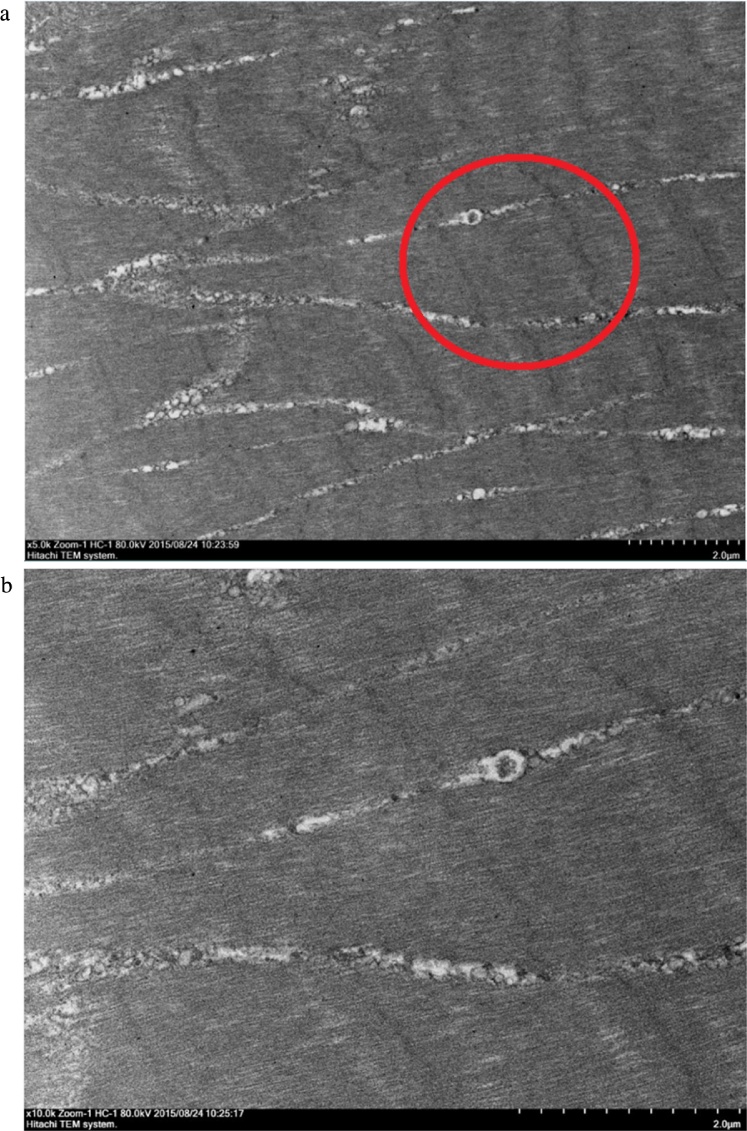
Transmission Electron Microscope (TEM) micrographs showing a longitudinal cross section of the muscle morphology of healthy, uninfected M. rosenbergii muscle tissues. The red circle indicates the area selected for a deeper look, focusing on fiber uniformity within the muscle bundles. Muscle fibers appear highly uniform, with the bundle structures well defined and orderly. The A band of the muscle tissue is also very clearly visible even at the lower resolution. A: TEM image showing healthy M. rosenbergii muscle fibers at a magnification of 5000×. (B) TEM image showing healthy M. rosenbergii muscle fibers at a magnification of 10000×.
Fig. 5.
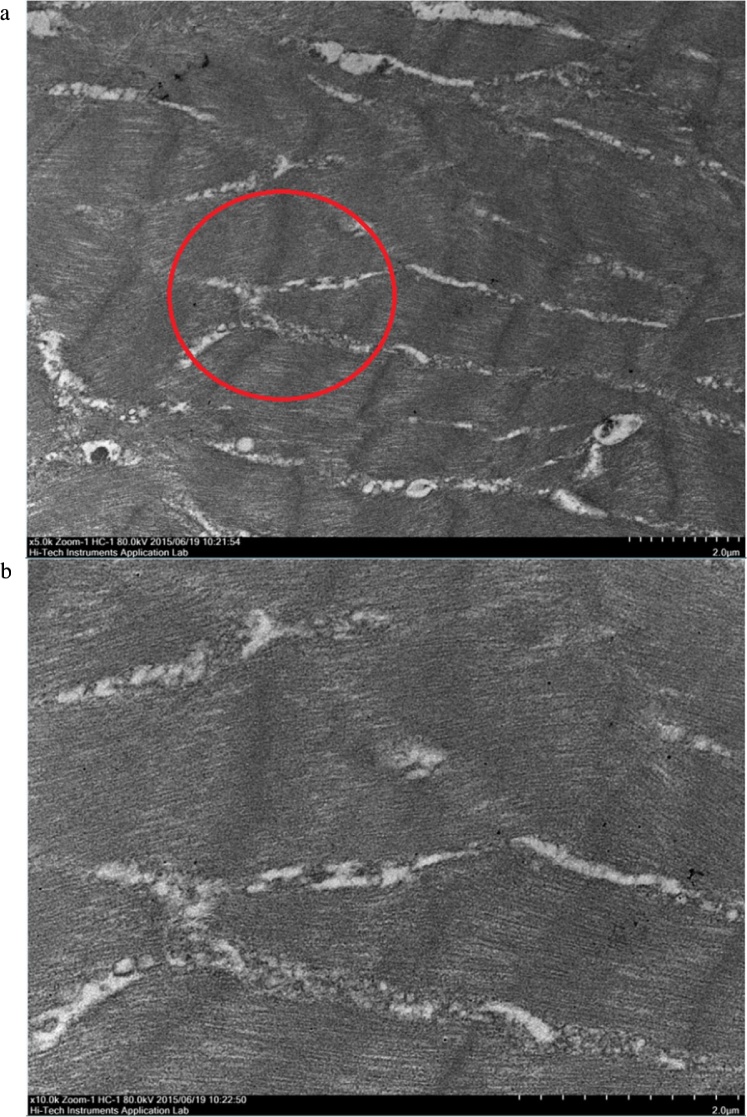
Transmission Electron Microscope (TEM) micrographs showing a longitudinal cross section of the muscle morphology of WSSV infected M. rosenbergii muscle tissues 24 hours post infection. The red circle indicates the area selected for a deeper look, focusing on muscle tissue fibers. Muscle fibers appear less defined compared to uninfected tissues. Although the tissue bands are visible, it is less orderly compared to uninfected tissues. (A) TEM image showing WSSV infected M. rosenbergii muscle fibers 24 hours post infection at a magnification of 5000×. (B) TEM image showing WSSV infected M. rosenbergii muscle fibers 24 hours post infection at a magnification of 10000×.
Fig. 6.
Transmission Electron Microscope (TEM) micrographs showing a longitudinal cross section of the muscle morphology of WSSV infected M. rosenbergii muscle tissues at 48 hours post infection. The red circle indicates the area selected for a deeper look, focusing on fiber uniformity with the muscle bundles. The A-band of the tissue appears distorted, reflecting visible distortions in muscle fibers. Arrangement of muscle bundles also appear to be distorted and visibly far less orderly compared to uninfected samples. (A) TEM image showing WSSV infected M. rosenbergii muscle fibers 48 hours post infection at a magnification of 5000×. (B) TEM image showing WSSV infected M. rosenbergii muscle fibers 48 hours post infection at a magnification of 10000×.
Fig. 7.
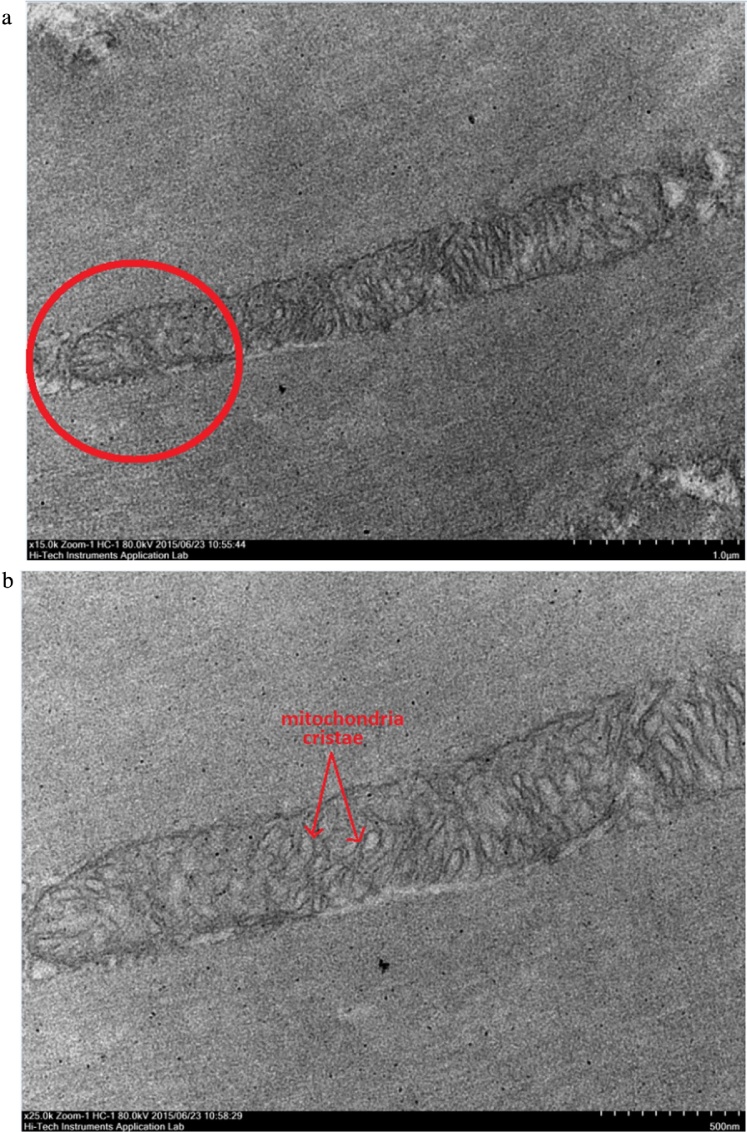
Transmission Electron Microscope (TEM) micrographs showing a mitochondrion organelle in a healthy M. rosenbergii muscle tissue. The red circle highlights the area selected to be observed at a higher magnification. Fig. 7B shows the close up image of the ultrastructure of healthy mitochondria cristae. (A) TEM image showing healthy M. rosenbergii muscle tissue mitochondrion organelle at a magnification of 15000×. (B) TEM image showing healthy M. rosenbergii muscle tissue mitochondrion organelle at a magnification of 25000×.
Fig. 8.
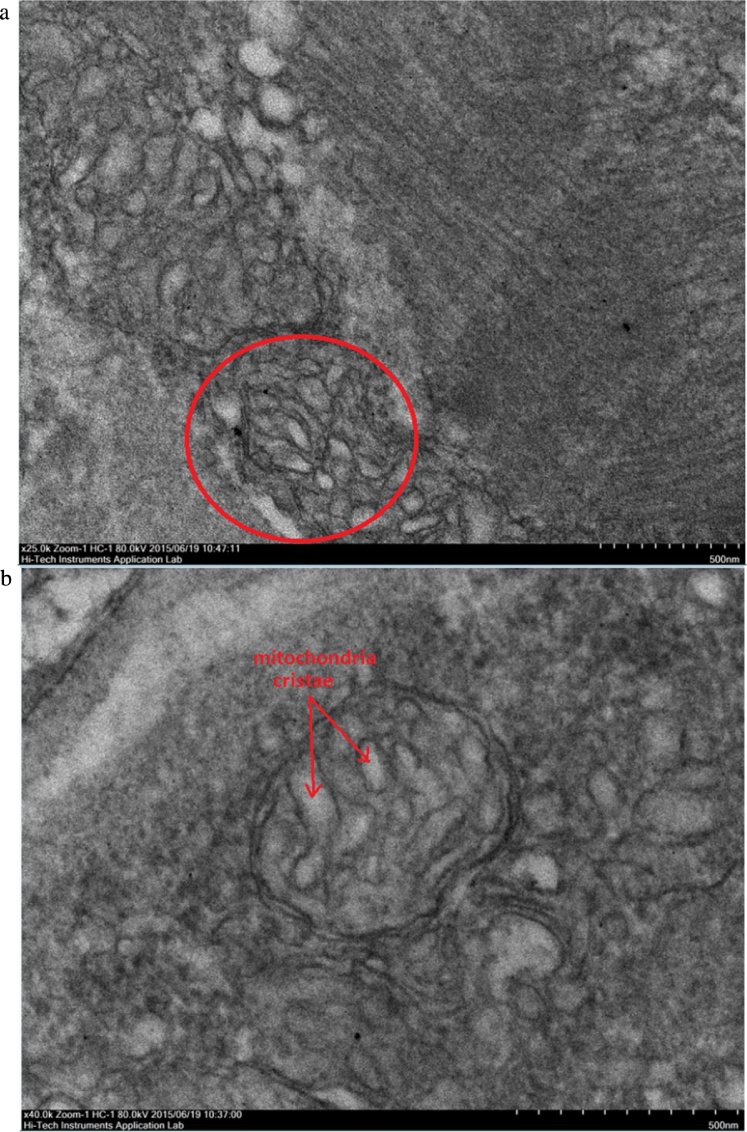
Transmission Electron Microscope (TEM) micrographs showing a mitochondrion organelle in WSSV infected M. rosenbergii muscle tissues at 24 hours post infection. The red circle highlights the area selected to be observed in higher magnification. (A) TEM image showing WSSV infected M. rosenbergii muscle tissue mitochondrion organelle 24 hours post infection at a magnification of 25000×. (B) TEM image showing WSSV infected M. rosenbergii muscle tissue mitochondrion organelle 24 hours post infection at a magnification of 40000×. This displayed the visibly swollen state of the mitochondria cristae.
Fig. 9.
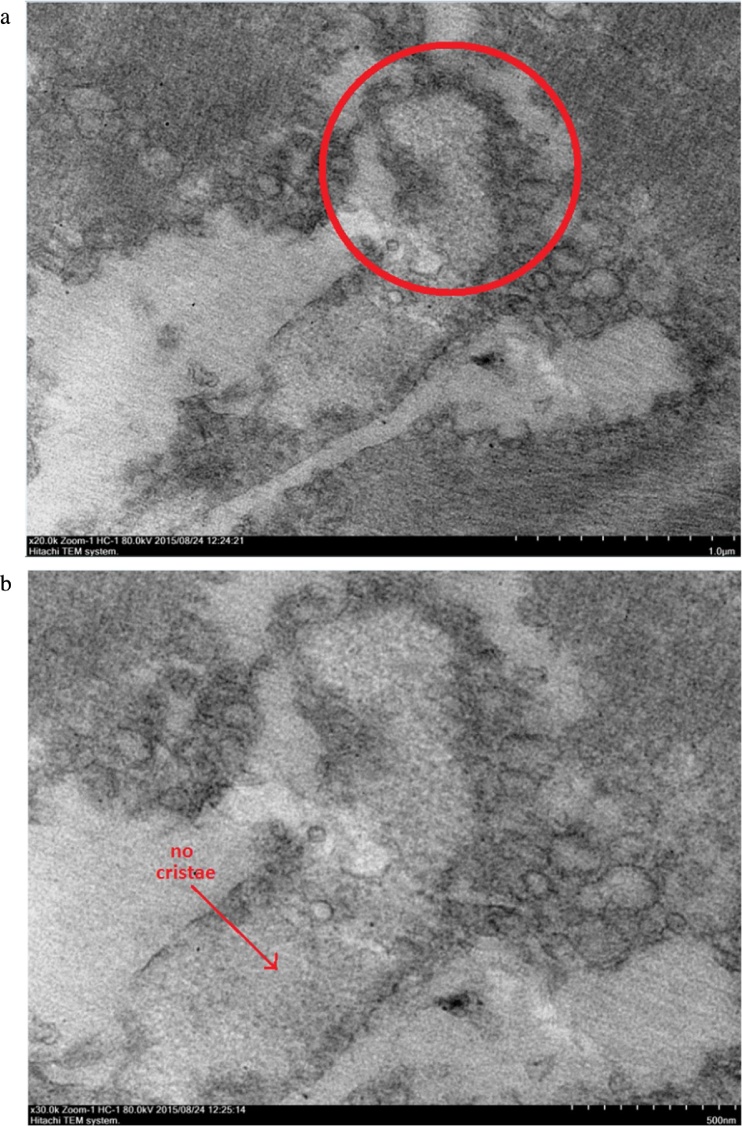
Transmission Electron Microscope (TEM) micrographs showing a mitochondrion organelle in WSSV infected M. rosenbergii muscle tissues at 48 hours post infection. The red circle highlights the area selected to be observed in higher magnification. (A) TEM image showing WSSV infected M. rosenbergii muscle tissue mitochondrion organelle 24 hours post infection at a magnification of 20000×. (B) TEM image showing WSSV infected M. rosenbergii muscle tissue mitochondrion organelle 24 hours post infection at a magnification of 30000×. This displayed the erupted state of the mitochondria cristae with no cristae and very few other ultrastructures remain inside.
The effect of dystrophin was determined through the conditions of the observed muscle fibers and muscle disk alignments, particularly the z-disk being the most affected muscle part by dystrophin. Disorders in the disk alignments and degraded states of muscle fibers were seen in the samples at 48 hours post infection, with noticeable breakages in the fiber arrangements. On the other hand, equal and orderly alignments were seen in the control samples.
Similarly, the mitochondrial organelles in the tissue samples of healthy M. rosenbergii were intact in the ultra-structural TEM images. Abnormal swellings of the cristae were observed in the mitochondria at 24 hours post infection when compared to healthy samples. Meanwhile, mitochondria cristae at samples of 48 hours post WSSV infection were absent, presumably damaged through excessive swelling.
4. Discussion
As an essential protein involved in muscular functioning, deficiency or loss of function of dystrophin can cause muscle wasting disorders and consequently cardiac or respiratory disabilities (Pradeep et al., 2012). A transcriptomic analysis by Rao et al. (2016) showed a down-regulation of dystrophin gene expression in white spot syndrome virus (WSSV) infected Macrobrachium rosenbergii samples compared to healthy control samples. Based on this experimental result, we hypothesised that WSSV affects both the mRNA expression of dystrophin and intracellular calcium concentrations, resulting in muscle weakness syndrome. Intracellular calcium concentration is also taken into consideration as one of the factor investigated due to its importance in normal muscular functioning.
Following up Rao et al.’s findings (2016), M. rosenbergii dystrophin (MrDys) sequences were retrieved from the NCBI Short Read Archives. Subsequently, corresponding primers were designed to conduct PCR amplification and subsequent sequencing in order to obtain the full M. rosenbergii dystrophin (MrDys) sequence. Using the obtained MrDys sequence as the template, the exact steps were repeated for obtaining the full P. monodon dystrophin (PmDys) sequence. The PmDys was necessary as it was a very closely related shrimp species dystrophin gene to MrDys and thus providing a deeper insight into the evolutionary relationships between the different dystrophin sequences. The MrDys and PmDys sequences obtained can then be utilized through the BLAST homology analysis which both showed high similarities to analogous dystrophin sequences from other invertebrate species in the phylum of Arthropoda, including Pogonomyrmex barbatus, Acyrthosiphon pisum and Athalia rosae.
Even though the MrDys (Supplementary Fig. 1a) and PmDys (Supplementary Fig. 2a) sequences obtained contain only partial coding sequence, the sequences discovered are sufficient as they already contain all of the four important protein domains of dystrophin conserved across many different species leaving only less significant parts upstream of Spectrin domain and downstream of ZZ superfamily domain left uncovered. In order to uncover such less significant portion of dystrophin sequences, a great amount of money, effort and time is required for conducting gene walking, for example, using RACE technique. However, the identification of the full coding sequence of dystrophin for both MrDys and PmDys will not contribute significantly to the current stage of the research, which involves mainly the construction of a simple but essential correlation for the functional role of dystrophin during WSSV infection. These partial dystrophin sequences can act as important foundation for understanding biochemical properties of dystrophin protein using techniques including dsRNA silencing, CRISPR silencing and point mutation analysis by other research groups and also increase citations of this manuscript when published. Thus, the partial dystrophin sequences are deemed to be adequate for the current stage of research and future genetic manipulation research.
Furthermore, a comparison was done at the protein domain level which successfully identified four distinct conserved domains in MrDys and PmDys, namely: the Spectrin, WW, EF Hand DMD-Like and ZZ superfamily domains (Jin et al., 2007). These four domains were important in connecting the intracellular cytoskeleton of actin to the intracellular matrix which was crucial for signalling, maintaining membrane stability and building muscle structure (Goldstein and McNally, 2010). The unique calponin domain found only in vertebrates may play an important role in causing differential dystrophin protein functioning between vertebrates and invertebrates.
Interestingly, our phylogenetic tree analysis, using the same thirteen dystrophin sequences, in the aspect of phyla comparison, pointed to MrDys being most closely related to Caenorhabditis elegans dystrophin, which was from phylum Nematoda (Fig. 1), with both species having highly similar proportions of conserved dystrophin sequence domains. Therefore, a further postulation can be made which was that the shrimp dystrophin gene became more evolutionarily diverged from the dystrophin gene of other phyla in the order of Nematoda, Arthropoda, Mollusca, Echinodermata and Chordata, in terms of genetic closeness, functionality and structural resemblances. The MrDys and PmDys showed the highest degree of similarity among the closely related species from an evolutionary point of view despite the difference in species habitat.
The comparison of dystrophin sequences across phyla was done at several levels, conducted through BLAST analysis, conserved domain analysis and phylogenetic analysis. These showed strong supports for the evolutionary conservation and divergence that occurred across species and phyla, especially between vertebrates and invertebrates. The studies of expression and functional roles of dystrophin gene during pathogenic infection using other species especially closely-related prawn species conducted by other researchers can be used as good references for inferring and supporting the expression changes and functional roles of dystrophin gene in M. rosenbergii during WSSV infection in this study.
The above findings were greatly supported by Roberts and Bobrow (1998), who found remarkable sequence conservation in dystrophin during the evolutionary process throughout the animal kingdom. Although the invertebrate dystrophin sequences studied were not completely identical to those of vertebrates, the dystrophin sequences of Cephalochordate amphioxus (lancelet), Asteroidea sp. (starfish), Pectinidae sp. (scallop), Drosophila melanogaster (fruit fly) and Caenorhabditis elegans (round worm) were shown to share a large number of characteristics with their vertebrate counterparts. These characteristics include the number of spectrin repetitions, EF Hand DMD-Like patterns and pairwise identical percentage of amino acid sequences as compared with the vertebrate dystrophin and dystrophin-like proteins (Roberts and Bobrow, 1998). These led Roberts and Bobrow to conclude that most metazoa possess sequences encoding a single highly conserved dystrophin-like protein in addition to a presumed distinct dystrobrevin, derived from an early duplication of an ancestral gene. It was also worth noting that multiple isoforms of dystrophin with shorter transcripts identified in sea urchin were found to bear significant resemblances to various forms of vertebrate dystrophin (Wang et al., 1998).
Another interesting aspect that emerged from our study was gene length. The gene length of MrDys was found to be broadly similar to that of some other dystrophin sequences. For instance, the short portion of the spectrin domain observed in MrDys had a similar relative length ratio to the spectrin domain found in C. elegans and D. melanogaster dystrophin (Supplementary Figure 4). On the other hand, dystrophin sequences retrieved from more complex organisms such as humans and dogs contained proportionately larger portions of the spectrin (SPEC) domain, with more repeats (Supplementary Figure 4). This evolutionary divergence was supported by the argument of Yan and Jeromin (2012) which stated that the expansion of the SPEC superfamily was due to the gene evolution from invertebrates to vertebrates: there are a total of 7 gene coding spectrin subunits in vertebrates, whereas there are only 3 such subunits in invertebrates. This helps to further explain why a noticeably shorter SPEC domain was observed in Macrobrachium rosenbergii.
A similar supporting evidence was the shorter gene coding regions of Heat Shock Protein 70 (HSP70) observed in invertebrates than vertebrates. Konstantopoulou et al. (1995) and Sültmann et al. (2000) reported that HSP70 for Drosophila auraria and Danio rerio were 633 amino acids and 659 amino acids respectively, compared to 702 amino acids for Homo sapiens (Fathallah et al., 1993).
While a smaller SPEC domain was observed in Macrobrachium rosenbergii, the other 3 domains in the species displayed similar sequence sizes to those of vertebrates. This implies a high level of structural conservation of the dystrophin gene between vertebrates and invertebrates. It is interesting to note that, despite the huge differences in dystrophin sequence lengths between mammals and arthropods (13 960 and 13 887 amino acids in humans and dogs respectively, against 414 amino acids in M. rosenbergii), the size of the EF-hand and ZZ conserved domains in MrDys were almost the same as their counterpart mammalian sequences.
A number of scientific research papers have suggested that WSSV infection contributes to muscle degeneration (Pradeep et al., 2012; Leu et al., 2007; Kou et al., 1998) which correlate with our findings that WSSV infected M. rosenbergii suffered muscle degeneration and consequent lethargy. The symptoms of muscle degeneration that were observed in WSSV infected M. rosenbergii included discoloration of the muscles and exoskeleton, lethargy and drastically reduced appetite.
In order to gain a deeper understanding into the relationship between WSSV and MrDys, we examined the WSSV copy numbers and mRNA expressions of MrDys in WSSV infected M. rosenbergii muscle tissues at different time intervals post-infection. The initial copy numbers of the virus from 0 hour to 24 hours post-infection did not change significantly as the viral infection was most probably being suppressed by the prawn innate immune system. Whereas for the mRNA expressions of MrDys, despite being less than two-fold change, it was considered as a significant fall of MrDys mRNA expression between 0 and 24 hours post-WSSV infection. This is supported by ANOVA analysis of the expressions which shows a significance level of P < 0.05 (Table 1). Along with this down-regulation of MrDys, a discoloration of muscles and exoskeleton were also observed. All these supported our presumption that the functional role of the dystrophin gene in preserving muscle integrity was affected by the down-regulation of MrDys (Shin et al., 2013).
Table 1.
Statistical ANOVA analysis of dystrophin-like gene expression in M. rosenbergii (MrDys) in response to WSSV infection by qPCR.
| ANOVA | |||||
|---|---|---|---|---|---|
| 2-dd(Ct) | |||||
| Sum of Squares | df | Mean Square | F | Sig. | |
| Between Groups | 5.598 | 6 | .933 | 11.213 | .000 |
| Within Groups | 1.165 | 14 | .083 | ||
| Total | 6.763 | 20 | |||
On the other hand, from 24 to 48 hours post infection, the WSSV copy number increased significantly and indicated the failure of cell’s innate immune system. This is similar with the reporting of Corteel et al. (2012) on the increase in the number of infected cells from the gills, stomach epithelium, cuticular epithelium and hematopoietic tissue of M. rosenbergii starting at 24-hours post WSSV infection. Thus, the increase in WSSV copy numbers may be postulated as the leading cause for the increased number of WSSV infected cells in the various organs (Corteel et al., 2012). Surprisingly, the mRNA expression levels of MrDys were up-regulated at 36 and 48 hours post infection, despite the increased copy numbers of WSSV virus during that time period. This up-regulation of MrDys was deduced as part of the prawn’s immune counter measures to prevent or reduce the muscle degeneration caused by WSSV infection and thus increasing the survivability of the WSSV infected M. rosenbergii (Goldstein and McNally, 2010). However, the observation of mortality for WSSV infected M. rosenbergii indicated the failure of such protective measure.
Any deficiencies in dystrophin and its associated proteins, dystroglycan and sarcoglycan, are generally accompanied by membrane damage and an alteration of calcium concentrations (Goonasekera et al., 2014; Mallouk et al., 2000). In our study, significant increase in intracellular calcium concentration [Ca2+]i was observed in muscle tissues at 6 hours post infection (Fig. 3). This sharp rise in [Ca2+]i occurred correspondingly to a decrease in MrDys expression as well as the loss of muscular integrity in the WSSV infected muscle tissues, suggesting a possible relationship between dystrophin expression and [Ca2+]i in the maintenance of muscular integrity. The reported functioning of dystrophin and its associated protein complexes in the stabilization of the cell membranes led to the prediction of membrane damage occurrence when in the absence of dystrophin. Then, this resulted in leakage or influx of calcium ions into the cells through the damaged membranes causing increased [Ca2+]i (Yeung et al., 2005; Miyake and McNeil, 2003; Mallouk et al., 2000; Petrof et al., 1993). Additionally, the activation of the Ca2+ permeable channels in dystrophin-deficient muscle membranes also supported our finding of increased [Ca2+]i upon the down-regulation of dystrophin gene (Allen et al., 2010). Another supporting postulation was the activation of calcium-activated enzyme, calpains in dystrophin-deficient muscle tissues, contributing to the increase of [Ca2+]i (Verburg et al., 2006).
Despite the most drastic decrease of dystrophin gene expression observed at 3 hours post WSSV infection, the most significant increase in [Ca2+]i, was actually observed at 6 hours post WSSV infection. It is deduced that most likely the downregulated dystrophin gene expression triggers a series of complex metabolic changes that eventually leads to the increase of [Ca2+]i,. However, certain amounts of transmission time required by such metabolic changes and also the possible existence of a threshold that maintains calcium ion influx are postulated to be the main factors causing the 3 hours gap between the most drastic changes of dystrophin gene expression and [Ca2+]i,. Taking such time gap into consideration, the second TEM observation is therefore conducted at 24 hours post infection instead of earlier 6 hours or 12 hours to provide sufficient time for the degeneration in muscle integrity to occur.
One contrary finding of this study was the stabilization of [Ca2+]i in M. rosenbergii muscle tissue despite the increase in MrDys expression levels after 24-hours post infection (Fig. 2, Fig. 3). As stated previously in the findings of Allen et al. in year 2010, decreased dystrophin expression level or functional dystrophin protein was associated with increased [Ca2+]i and vice versa. However, in this finding, [Ca2+]i level did not decrease after 24-hours post WSSV infection despite the increase in MrDys expression level. Thus, this contrary finding led to the postulation that the maintenance of [Ca2+]i level was due to the same amount of functional MrDys protein. This is supported by another assumption of MrDys protein degradation due to up-regulation of chrymotrypsin which subsequently led to the loss of muscle integrity. According to Xue et al. (2013), chymotrypsin was up-regulated 14 times in a WSSV infected shrimp compared to an uninfected specimen. They also reported reduced shrimp mortality and WSSV copy numbers in chrymotrypsin knock-out shrimps. In addition, Yoshida et al. (1992) demonstrated that dystrophin contained several proteinase cleavage sites and became several polypeptide fragments upon alpha chrymotrypsin digestion (Yoshida et al., 1992). Thus, chrymotrypsin was postulated to be up-regulated upon WSSV infection and consequently resulting in the increased proteolytic degraded dystrophin. Therefore, this explained the maintenance of [Ca2+]i level and continued loss of muscular integrity due to MrDys protein degradation despite the up-regulation of the MrDys gene expression.
Transmission Electron Microscope (TEM) observations were carried out based on two different aspects of the muscle tissues, the longitudinal cross section layout of muscle tissues, and the mitochondria found in between the muscle fiber bundles. The longitudinal cross section was selected as it gave a very profound observation on the layout and uniformity of the muscle bundles. The muscle bundles were compared based on the alignment of the bundles, inter-bundle space and general appearance of organelles. Meanwhile, observations of the mitochondria ultra-structure focused on the structure of cristae and membranes within the organelle. The TEM images of muscular fibres were displayed progressively in the order of normal muscle (Fig. 4), 24 hours post-WSSV infection muscle (Fig. 5), 48 hours post-WSSV infection muscle (Fig. 6). Another view was taken into the mitochondria condition under normal (Fig. 7), 24 hours post-WSSV infection (Fig. 8) and 48 hours post-WSSV infection (Fig. 9). These TEM image comparisons showed clearly the progressive muscle deterioration during WSSV infection in M. rosenbergii. Two panels, A and B, were provided for every TEM image. Panel A showed a more general view with less magnification whereas panel B showed the interested region in higher magnification.
In the longitudinal cross section comparison, two different magnifications, 5000× and 10000×, were used to compare the muscle fibers and bundles. The micrographs of longitudinal cross section of healthy M. rosenbergii muscle tissues were shown in Fig. 4, showing uniformed and aligned muscle fibers as well as highly distinct well preserved bands. The micrographs of WSSV infected M. rosenbergii muscle tissues 24 hours post infection (Fig. 5) showed less distinct muscle bands and visible disturbances in its alignments especially observable under increased magnification (Fig. 5B). A further decrease in the uniformity and alignment of muscle bundles, bands and fibers was observed for micrographs of 48 hours post WSSV infection (Fig. 6A) accompanied with high level of muscle fiber disorder at a magnification of 10000× (Fig. 6B).
This progressive deformation of muscle fibers concurs with the earlier discussed theory regarding the correlation between muscular changes caused by WSSV infection and dystrophin gene expression in M. rosenbergii. The gradual worsening disorder conditions of the muscle tissues from 0 hours to 24 hours and finally to 48 hours WSSV post infection were due to the WSSV induced decrease in dystrophin expression. The elevated protein cleavage of dystrophin during WSSV infection was also one of the contributing factor as discussed previously.
Meanwhile, progressive muscular damages due to WSSV infection were also observable in the mitochondria structures of M. rosenbergii. The mitochondrial cristae in healthy M. rosenbergii tissues appeared well defined and organized within the organelle membrane (Fig. 7). Swelling of the cristae was observed in muscle tissues 24 hours post WSSV infection showing larger and rounded cristae structures (Fig. 8). The excessive cristae swellings at 24 hours post infection actually functioned as indicator for the eventual cristae bursting which caused the disappearance of cristae in muscle tissues 48 hours post WSSV infection (Fig. 9). This supported the inference concerning the swelling and eventual bursting of the cristae structure under the effect of WSSV infection which was vital for the severe impairment of the muscle tissue’s ability to generate and utilize energy efficiently. This was shown by the blatant muscle weaknesses and lethargy observed in infected M. rosenbergii prawn samples.
5. Conclusion
After taking a deeper insight into the evolutionary relationships between dystrophin sequences from different phyla, the functioning of dystrophin in response to WSSV infection was then investigated. Importantly, our results demonstrated for the first time the relationship between the dystrophin gene and intracellular calcium concentrations in Macrobrachium rosenbergii infected by WSSV. Specifically, we observed mRNA dystrophin expression and intracellular calcium concentration changes in WSSV infected M. rosenbergii over a range of time intervals. The degree of changes in the muscle integrity in response to altered dystrophin expression levels during WSSV infection was shown clearly in the TEM images provided. Overall, our results highlighted the correlation between mRNA expression of MrDys and [Ca2+]i upon WSSV infection, and the consequent effects on muscle integrity.
This study involves the muscle degeneration triggered by downregulated dystrophin gene expression in prawn during WSSV infection. A simple correlative relationship was established between MrDys gene expression, [Ca2+]i and muscle integrity in M. rosenbergii. Cascade reactions are usually involved in signalling or metabolic activities and result in complex disease-related cause and effect relationships between different genes and proteins. Thus, the strong relationship can only be confirmed after the identification of all other genes involved in the muscle deterioration during WSSV infection through further studies.
The dominant functional role of dystrophin in maintaining muscle integrity of M. rosenbergii and the major effects of its reduced expression can be confirmed by including other genes that are related to muscular dystrophies, for example, myostatin, myosin heavy chain, dystroglycan, sarcoglycan, and tropomyosin as subjects of study as well. These dystrophin-related genes can be used as good positive or negative controls for comparisons after obtaining their expression changes during WSSV infection.
A research conducted previously by Sarasvathi in year 2016 can be used as supportive evidence for the functional role of MrDys. The research was focused on the characterization of myostatin gene in M. rosenbergii, which is an important muscle-related gene. This M. rosenbergii myostatin gene was named MrMSTN. Subsequent verification of the functional role of MrMSTN was conducted through dsRNA silencing, gene expression analysis and histological analysis. The primary role of MrMSTN as a negative growth regulator was verified. In addition, the correlation studies of MrMSTN also showed its secondary function in the expression alteration of other genes affected by the MrMSTN silencing. This was shown by the downregulation of myosin heavy chain gene expression. Dystrophin gene expression in M. rosenbergii was downregulated as well during the silencing. However, tropomyosin gene expression was upregulated during MrMSTN silencing. This showed the complex relationship between the muscle-related genes by which expression of one affects the others significantly either positively or negatively.
The postulated role of MrDys in the maintenance of muscle integrity can be further supported by the histological analysis of M. rosenbergii muscle tissue with silenced MrMSTN. The downregulated MrDys gene expression associated with silenced MrMSTN can be deduced to lead to muscle degeneration. This consequently triggers the muscle tissue proliferation and regeneration observed in histological analysis during 7 dpi (days post infection) and 14 dpi to recover from the damage. Therefore, the inclusion of other muscle-related genes in the WSSV challenge was deemed unnecessary at the current stage of research as the relationship between the genes was already proven during previous study. Nevertheless, this should be taken into consideration in subsequent experiment designs.
A suitable approach to be taken for the verification of the functional role and immune significance of dystrophin gene expression during WSSV infection would be dystrophin gene knockdown through dsRNA silencing technique and the expression study of the silenced gene during WSSV infection. This approach can also help to strengthen the postulated simple correlation between dystrophin gene expression, [Ca2+]i and the muscular deterioration during WSSV infection. However, due to several unavoidable limitations faced in terms of grant and time, such validation approach can only be retained for future research.
The general work flow for the follow-up research can be divided into three main stages. The first stage being the identification of the full-length dystrophin coding sequences of different shrimp species of interest including M. rosenbergii. Possible long form or isoform dystrophin sequences can also be discovered in the process. This stage is already in progress being conducted by another interested researcher under the same research group of the author. After the completion of first stage, stringent experiment design will be carried out for the dsRNA silencing of dystrophin gene, its expression study and TEM observation under WSSV challenged condition while considering the inclusion of other muscle-related genes as positive and negative controls for validating the influence of dystrophin gene expression on [Ca2+]i and muscular integrity. Lastly, after the gene expression level, the research can be taken another step further towards protein studies.
Declarations
Author contribution statement
Anees Fathima Noor, Tze Chiew Christie Soo: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Farhana Mohd Ghani, Zee Hong Goh, Li Teng Khoo: Analyzed and interpreted the data; Wrote the paper.
Subha Bhassu: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Competing interest statement
The authors declare no conflict of interest.
Funding statement
This work was funded by the High Impact Research Centre, University of Malaya (UM.C/625/HIR/MOHE/AS01). The funders had no role in the experimental design, data collection, analysis or the preparation of the manuscript.
Additional information
Data associated with this study has been deposited at the GenBank database under the accession number KU198993.
Appendix A. Supplementary data
References
- Allen D.G., Gervasio O.L., Yeung E.W., Whitehead N.P. Calcium and the damage pathways in muscular dystrophy. Can. J. Physiol. Pharmacol. 2010;88(2):83–91. doi: 10.1139/Y09-058. [DOI] [PubMed] [Google Scholar]
- Bateman K.S., Tew I., French C., Hicks R.J., Martin P., Munro J., Stentiford G.D. Susceptibility to infection and pathogenicity of White Spot Disease (WSD) in non-model crustacean host taxa from temperate regions. J. Invertebrate Pathol. 2012;110(3):340–351. doi: 10.1016/j.jip.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Corteel M., Lima P.D.D.R., José J., Tuan V.V., Khuong T.V., Wille M., Alday-Sanz V., Pensaert M., Sorgeloos P., Nauwynck H. Susceptibility of juvenile Macrobrachium rosenbergii to different doses of high and low virulence strains of white spot syndrome virus (WSSV) Dis. Aquat. Organ. 2012;100(3):211–218. doi: 10.3354/dao02496. [DOI] [PubMed] [Google Scholar]
- Dhar A.K., Bowers R.M., Licon K.S., Veazey G., Read B. Validation of reference genes for quantitative measurement of immune gene expression in shrimp. Mol. Immunol. 2009;46(8–9):1688–1695. doi: 10.1016/j.molimm.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Fathallah D.M., Cherif D., Dellagi K., Arnaout M.A. Molecular cloning of a novel human hsp70 from a B cell line and its assignment to chromosome 5. J. Immunol. 1993;151(2):810–813. [PubMed] [Google Scholar]
- Goldstein J.A., McNally E.M. Mechanisms of muscle weakness in muscular dystrophy. J. Gen. Physiol. 2010;136(1):29–34. doi: 10.1085/jgp.201010436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonasekera S.A., Davis J., Kwong J.Q., Accornero F., Wei-LaPierre L., Sargent M.A., Dirksen R.T., Molkentin J.D. Enhanced Ca(2)(+) influx from STIM1-Orai1 induces muscle pathology in mouse models of muscular dystrophy. Hum. Mol. Genet. 2014;23(14):3706–3715. doi: 10.1093/hmg/ddu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed A.S., Balasubramanian G., Musthaq S.S., Yoganandhan K. Experimental infection of twenty species of Indian marine crabs with white spot syndrome virus (WSSV) Dis. Aquat. Organ. 2003;57(1/2):157–161. doi: 10.3354/dao057157. [DOI] [PubMed] [Google Scholar]
- Jin H., Tan S., Hermanowski J., Böhm S., Pacheco S., McCauley J.M., Greener M.J., Hinits Y., Hughes S.M., Sharpe P.T., Roberts R.G. The dystrotelin, dystrophin and dystrobrevin superfamily: new paralogues and old isoforms. BMC Genomics. 2007;8(1) doi: 10.1186/1471-2164-8-19. p.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantopoulou I., Ouzounis C.A., Drosopoulou E., Yiangou M., Sideras P., Sander C., Scouras Z.G. A Drosophila hsp70 gene contains long, antiparallel, coupled open reading frames (LAC ORFs) conserved in homologous loci. J. Mol. Evol. 1995;41(4):414–420. doi: 10.1007/BF00160312. [DOI] [PubMed] [Google Scholar]
- Kou G.H., Peng S.E., Chiu Y.L., Lo C.F. Tissue distribution of white spot syndrome virus (WSSV) in shrimp and crabs. Adv. Shrimp Biotechnol. 1998:267–271. [Google Scholar]
- Leu J.H., Chang C.C., Wu J.L., Hsu C.W., Hirono I., Aoki T., Juan H.F., Lo C.F., Kou G.H., Huang H.C. Comparative analysis of differentially expressed genes in normal and white spot syndrome virus infected Penaeus monodon. BMC Genomics. 2007;8(1):1–14. doi: 10.1186/1471-2164-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mallouk N., Jacquemond V., Allard B. Elevated subsarcolemmal Ca2+ in mdx mouse skeletal muscle fibers detected with Ca2+-activated K+ channels. Proc. Natl. Acad. Sci. 2000;97(9):4950–4955. doi: 10.1073/pnas.97.9.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Cano F., Sánchez-Paz A. Development and validation of a quantitative real-time polymerase chain assay for universal detection of the White Spot Syndrome Virus in marine crustaceans. Virol. J. 2013;10(1):1. doi: 10.1186/1743-422X-10-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K., McNeil P.L. Mechanical injury and repair of cells. Crit. Care Med. 2003;31(8 Suppl):S496–501. doi: 10.1097/01.CCM.0000081432.72812.16. [DOI] [PubMed] [Google Scholar]
- Muntoni F., Torelli S., Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2(12):731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. U. S. A. 1993;90(8):3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeep B., Rai P., Mohan S.A., Shekhar M.S., Karunasagar I. Biology, host range, pathogenesis and diagnosis of white spot syndrome virus. Indian J. Virol. 2012;23(2):161–174. doi: 10.1007/s13337-012-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran R.B., Rajendran K.V., Jung S.J., Oh M.J. Experimental susceptibility of different life‐stages of the giant freshwater prawn, Macrobrachium rosenbergii (de Man), to white spot syndrome virus (WSSV) J. Fish Dis. 2002;25(4):201–207. [Google Scholar]
- Rao R., Bhassu S., Bing R.Z., Alinejad T., Hassan S.S., Wang J. A transcriptome study on Macrobrachium rosenbergii hepatopancreas experimentally challenged with White Spot Syndrome Virus (WSSV) J. Invertebrate Pathol. 2016 doi: 10.1016/j.jip.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Roberts R.G., Bobrow M. Dystrophins in vertebrates and invertebrates. Hum. Mol. Genet. 1998;7(4):589–595. doi: 10.1093/hmg/7.4.589. [DOI] [PubMed] [Google Scholar]
- Ruegg U.T., Gillis J.-M. Calcium homeostasis in dystrophic muscle. Trends Pharmacol. Sci. 1999;20(9):351–352. doi: 10.1016/s0165-6147(99)01377-2. [DOI] [PubMed] [Google Scholar]
- Rybakova I.N., Patel J.R., Ervasti J.M. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 2000;150(5):1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasvathi E. University of Malaya; Kuala Lumpur, Malaysia: 2016. Myostatin-like Gene: A Potential Target for Gene Silencing in Giant Freshwater Prawn, Macrobrachium rosenbergii (Doctoral Dissertation) [Google Scholar]
- Shin J., Tajrishi M.M., Ogura Y., Kumar A. Wasting mechanisms in muscular dystrophy. Int. J. Biochem. Cell Biol. 2013;45(10):2266–2279. doi: 10.1016/j.biocel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sültmann H., Sato A., Murray B.W., Takezaki N., Geisler R., Rauch G.J., Klein J. Conservation of Mhc class III region synteny between zebrafish and human as determined by radiation hybrid mapping. J. Immunol. 2000;165(12):6984–6993. doi: 10.4049/jimmunol.165.12.6984. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verburg E., Dutka T.L., Lamb G.D. Long-lasting muscle fatigue: partial disruption of excitation-contraction coupling by elevated cytosolic Ca2+ concentration during contractions. Am. J. Physiol. 2006;290(4):C1199–C1208. doi: 10.1152/ajpcell.00469.2005. [DOI] [PubMed] [Google Scholar]
- Wang J., Pansky A., Venuti J.M., Yaffe D., Nudel U. A sea urchin gene encoding dystrophin-related proteins. Hum. Mol. Genet. 1998;7(4):581–588. doi: 10.1093/hmg/7.4.581. [DOI] [PubMed] [Google Scholar]
- Xue S., Yang W., Sun J. Role of chymotrypsin-like serine proteinase in white spot syndrome virus infection in Fenneropenaeus chinensis. Fish Shellfish Immunol. 2013;34(2):403–409. doi: 10.1016/j.fsi.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Yan X.X., Jeromin A. Spectrin breakdown products (SBDPs) as potential biomarkers for neurodegenerative diseases. Curr. Transl. Geriatr. Exp. Gerontol. Rep. 2012;1(2):85–93. doi: 10.1007/s13670-012-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Suzuki A., Shimizu T., Ozawa E. Proteinase-sensitive sites on isolated rabbit dystrophin. J. Biochem. 1992;112(4):433–439. doi: 10.1093/oxfordjournals.jbchem.a123918. [DOI] [PubMed] [Google Scholar]
- Yeung E.W., Whitehead N.P., Suchyna T.M., Gottlieb P.A., Sachs F., Allen D.G. Effects of stretch-activated channel blockers on [Ca2+]i and muscle damage in the mdx mouse. J. Physiol. 2005;562(Pt 2):367–368. doi: 10.1113/jphysiol.2004.075275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.