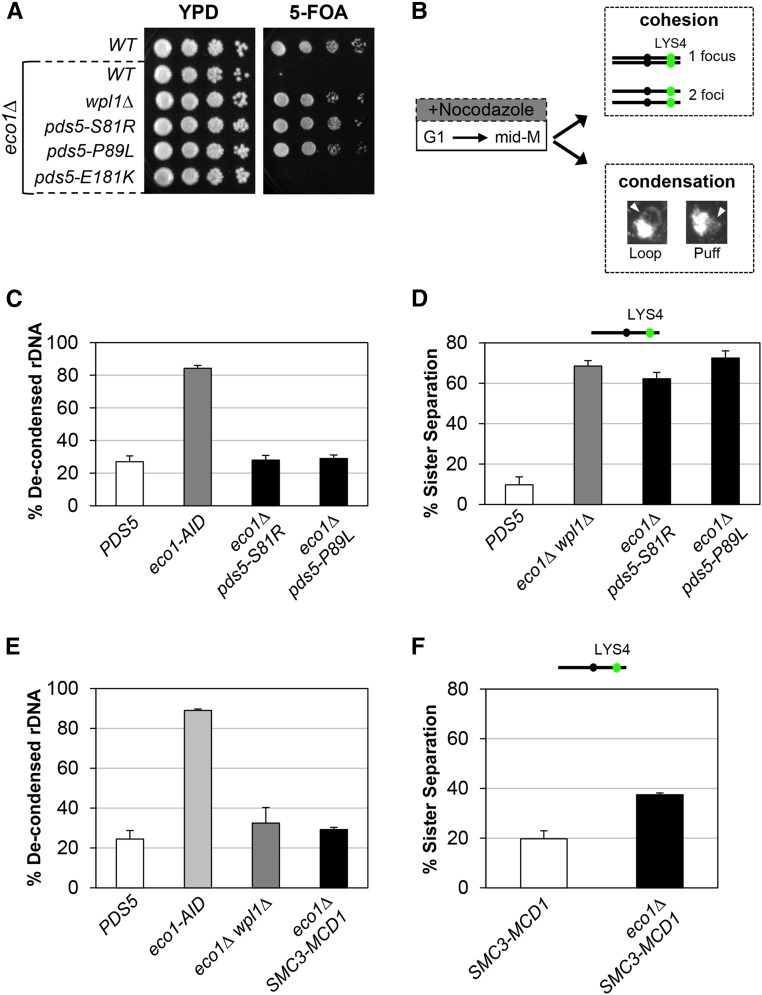
Figure 3.
pds5 N-terminal mutants and SMC3-MCD1 fusion suppress inviability of eco1∆ through restoration of condensation. (A) Plasmid shuffle assay to assess viability of pds5 N-terminal mutants in the eco1∆ background. Plasmid pBS1030 (ECO1 CEN URA3) is present in haploid wild-type (WT) (VG3349-1B), eco1∆ (VG3499-1B), eco1∆ wpl1∆ (VG3503 #4), eco1∆ pds5-S81R (MSB138-1K), eco1∆ pds5-P89L (MSB139-2J), and eco1∆ pds5-E181K (MSB147-1A) strains. Cells were grown to saturation in YPD media at 23°, plated at 10-fold serial dilutions on YPD or 5-FOA media, and then incubated for 3 days at 23°. 5-FOA selects for loss of pBS1030 (ECO1 CEN URA3). (B) Schematic of time course and analysis of cohesion and condensation. Cells were synchronously arrested in mid-M-phase as described in the Materials and Methods. Cells were processed for cohesion analysis of LacI-GFP at the CEN-distal LYS4 locus and CEN-proximal TRP1, and for condensation by FISH methodology (Materials and Methods). (C) pds5-S81R and pds5-P89L restore condensation in eco1∆ cells. PDS5 (VG3349-1B), eco1-AID (VG3633-2D), eco1∆ pds5-S81R (MSB138-1K), and eco1∆ pds5-P89L (MSB139-2J) were arrested in G1 using α-factor, then synchronously arrested in mid-M-phase using nocodazole as described in the Materials and Methods. From G1 through mid-M-phase, 500 µM auxin was present in the media of the eco1-AID strain. Cells were fixed and processed for FISH (Materials and Methods). Chromosome condensation was assessed by morphology of the rDNA locus and cells were scored for condensed rDNA (loops) and defective condensation (puffs). The percentage of cells with defective rDNA condensation (decondensed) is plotted. (D) pds5-S81R eco1∆ and pds5-P89L eco1∆ double-mutants have a dramatic defect on cohesion. PDS5 (VG3349-1B), eco1∆ wpl1∆ (VG3503 #4), eco1∆ pds5-S81R (MSB138-1K), and eco1∆ pds5-P89L (MSB139-2J) cells were synchronously arrested in mid-M-phase using nocodazole (Materials and Methods). Cells were scored for cohesion (one GFP focus) and loss of cohesion (two GFP foci; sister separation) at the CEN-distal LYS4 locus. The percentage of cells lacking cohesion (sister separation) is shown. (E) SMC3-MCD1 fusion promotes condensation in eco1∆ cells. WT (VG3349-1B), eco1-AID (VG3633-2D), eco1∆ wpl1∆ (VG3502 #A), and eco1∆ SMC3-MCD1 (MSB249-3A) were synchronously arrested in mid-M-phase (Materials and Methods). From G1 through mid-M-phase, 500 µM auxin was present in the media of the eco1-AID strain. Cells were fixed and processed for FISH to assess rDNA condensation (loops) and defective condensation (puffs), as described in (C). The percentage of cells with defective rDNA condensation (decondensed) is plotted. (F) SMC3-MCD1 fusion partially restores cohesion to eco1∆ cells. SMC3-MCD1 (VG3940-2D) and eco1∆ SMC3-MCD1 (MSB249-3A) were synchronously arrested in mid-M-phase using nocodazole (Materials and Methods). Cells were scored for cohesion (one GFP focus) and loss of cohesion (two GFP foci; sister separation) at the CEN-distal LYS4 locus as described in (B). The percentage of cells lacking cohesion (sister separation) is shown. The strains in this panel and in Figure 4C were analyzed for cohesion loss in the same experiment. The data were separated for clarity of presentation, and the SMC3-MCD1 cohesin data are presented here and in Figure 4C.