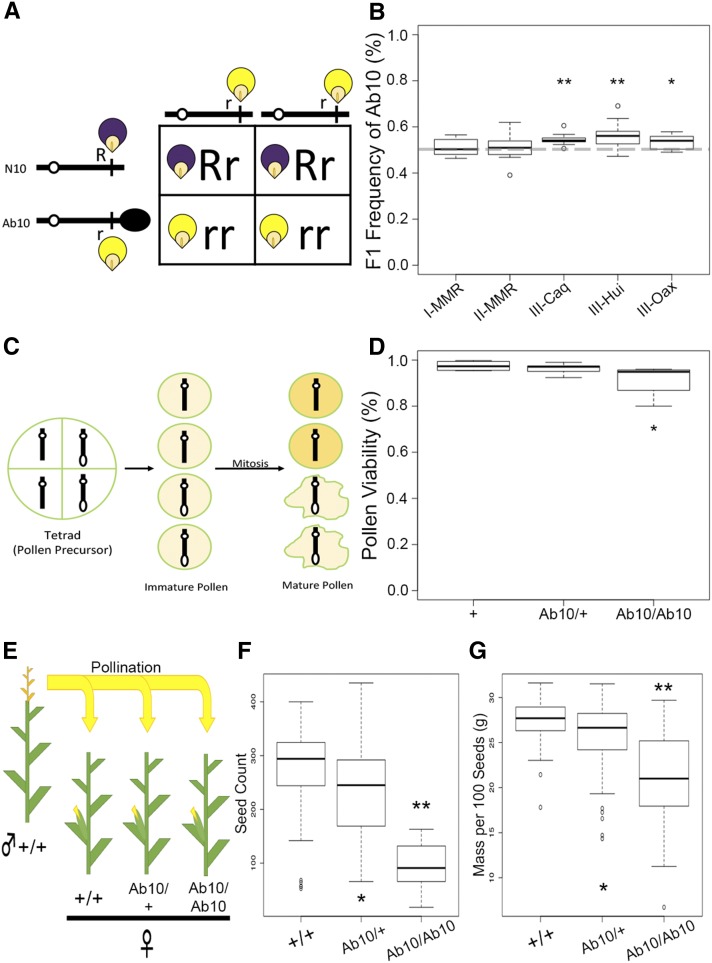
Figure 3.
Phenotypic consequences of Ab10-I-MMR. (A) Punnett square diagramming the testcrossing scheme used to measure inheritance of Ab10. In this case, the Ab10 haplotype is linked to the recessive allele of r1, resulting in yellow kernels. The dominant allele R1 is linked to N10 and conditions purple kernels. (B) Quantification of the transmission rates of five Ab10 haplotypes as a male parent. None of the medians were lower than 50%. Transmission of three of the haplotypes was significantly higher than expected as determined by Fisher’s Exact Test (* P < 0.01 and ** P < 0.0001). (C) Pollen development involves two rounds of mitosis in the haploid phase, where Ab10 could have negative phenotypic consequences (here shown as crinkled, dead pollen). (D) Pollen viability in different Ab10-I-MMR genotypes. Pollen viability in N10/N10 homozygotes ( = 0.97) and Ab10/N10 heterozygotes ( = 0.96) were not significantly different, whereas Ab10/Ab10 homozygotes had significantly lower pollen viability ( = 0.91) than both N10/N10 and Ab10/N10 (P = 0.049 and 0.009, respectively). (E) Crossing scheme for measuring the effects of Ab10 on seed count and mass. All genotypes were in the B73 background. (F) Seed count for Ab10/Ab10 homozygotes ( = 95) and Ab10/N10 heterozygotes ( = 233) was significantly reduced compared to N10/N10 wild-types ( = 271). Statistical significance is shown (* P < 0.05 and ** P < 0.001) as measured by Student’s t-test. (G) Mass per 100 seeds in Ab10 homozygotes ( = 20.8 g) and Ab10 heterozygotes ( = 25.7 g) is significantly reduced compared to N10 wild-types ( = 27.3 g). Ab10, abnormal chromosome 10; N10, normal chromosome 10.