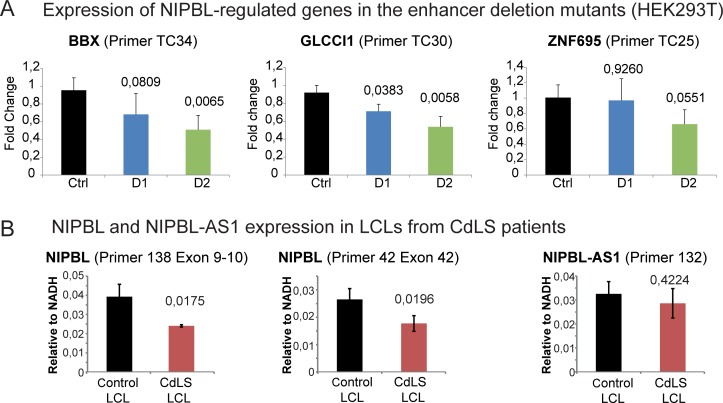
Fig 5. Implications for CdLS.
A) Transcript levels of the genes BBX, GLCCI1 and ZNF695 that were described as dysregulated genes in CdLS [20] and previously confirmed as NIPBL-dependent genes with NIPBL binding sites at the promoter [8] were analysed in the different enhancer deletion clones D1 and D2 (mean n = 5 for D1 and n = 4 for D2, error bars +/- s.d., p-values determined with t-Test, the transcript levels of the individual clones are shown in S7 Fig). B) Average transcript levels of NIPBL and NIPBL-AS1 in lymphoblastoid cell lines (LCLs) derived from CdLS patients and controls. The details of the four LCL controls and three CdLS LCLs as well as the individual transcript levels are shown in S8 Fig and in [8,20]. Two primer pairs for NIPBL and one for NIPBL-AS1 were used. Transcript levels were normalized against the housekeeping gene NADH (mean n = 4 for control LCLs and n = 3 for CdLS LCLs, error bars +/- s.d., p-values determined with t-Test).