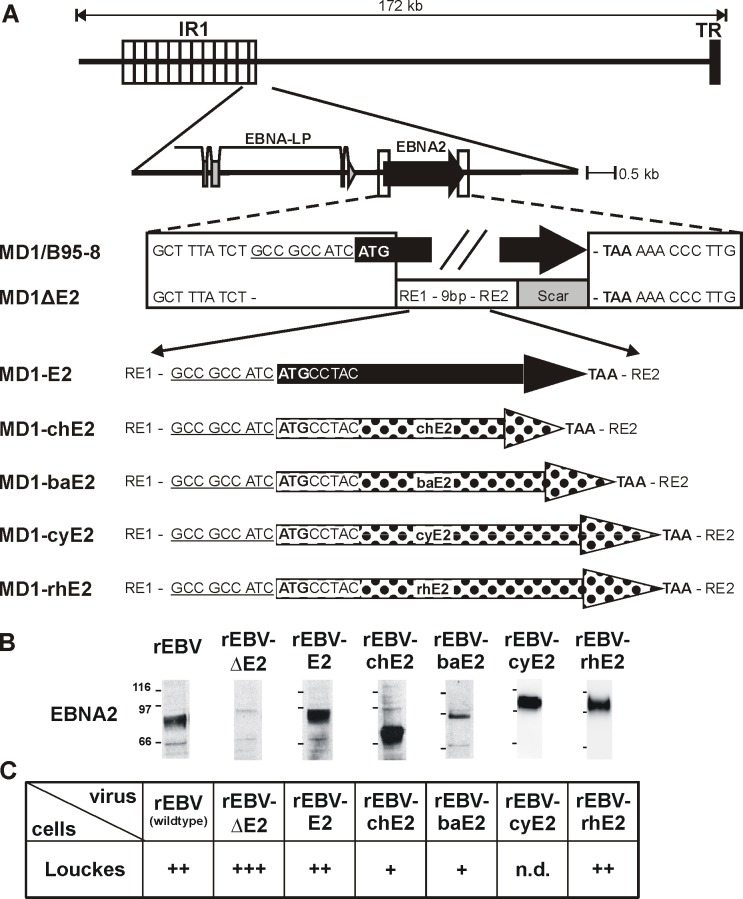
Fig 1. Recombinant EBVs expressing NHP EBNA2 proteins infect human B cells.
A) Relative location of the EBNA2 gene locus downstream of the major Internal Repeat (IR1) within the EBV genome and detailed description of the flanking regions within the EBV BAC (MD1). The native flanking sequences and EBNA2 open reading frame (black arrow) are shown in the wild type EBV BAC derived from B95-8 (MD1/B95-5). The EBNA2 deleted BAC (MD1ΔE2) contains the linker region from the shuttle plasmid containing two restriction enzyme sites (RE1 and 2) and a downstream scar sequence as a result of the cloning procedure. Recombinant EBVs carrying EBV E2 (MD1-E2, black arrow), chimpanzee, baboon, cynomolgus or rhesus LCV EBNA2 (MD1-chE2, -baE2, -cyE2, -rhE2; polka dots) were cloned by recombining similar DNA fragments from shuttle plasmids containing each EBNA2 species. In this manner all rEBV contained an EBNA2 species with identical upstream and downstream sequences. B) Production of rEBVs was confirmed by infection of EBV-negative Louckes cells and detection of EBNA2. The monoclonal antibody against EBNA2 (PE2) was used to detect LCV EBNA2 expression by immunoprecipitation and subsequent immunoblotting from cell lysates prepared 2 days after infection with equal volumes of rEBV. C) Relative infectious titers of rEBV supernatants were determined by infection of EBV-negative Louckes cells and puromycin selection. Louckes cells were infected with equal volumes of rEBV, distributed into 50 microtiter wells, and selected for rEBV infection using puromycin. After 3–4 weeks the number of wells with cell growth was counted, and average results for at least 2 independent supernatants are shown. +: 1–20 wells with growth/50 total wells, ++: 21-44/50, +++: 45-50/50, n.d.: not done.