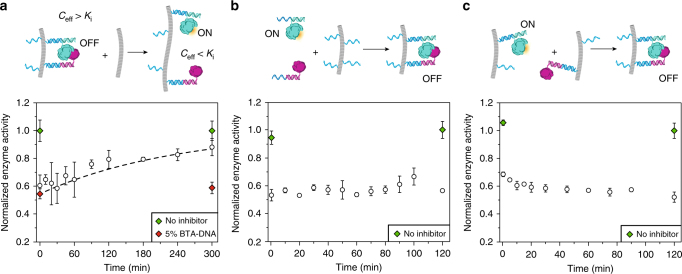
Fig. 4.
Characterization of the dynamics of enzyme-inhibitor recruitment on the supramolecular polymer. a Enzyme activity as a function of time after the addition of pre-assembled polymers containing 100% BTA-3OH (9.5 μL, 1 mM) to polymers composed of 5% BTA-DNA (0.5 μM BTA-DNA, 9.5 μM BTA-3OH), 20 nM RE and RI, 1 nM β-lactamase and 10 nM BLIP (total volume = 40.5 μL), yielding a final BTA-DNA fraction of 0.25% (0.5 μM BTA-DNA, 199.5 μM BTA-3OH, final total volume = 50 μL). The dashed line represents the fitting of a single exponential with fixed values at t = 0 of the activity in the 5% BTA-DNA reference sample and at t = ∞ of the activity in the reference sample omitting BLIP yielding t 1/2 = ~3 h. The green diamonds represent the activity of a sample without inhibitor. The red diamonds represent the activity of the system containing 5% BTA-DNA, which was not diluted by addition of 100% BTA-3OH. b Kinetics of enzyme-inhibitor complex formation upon recruitment to the DNA-decorated polymers. Enzyme activity was measured at different time-point after adding the BTA polymers to a mixture of β-lactamase, BLIP RE and RI. Experiments were performed using 25% BTA-DNA (0.5 μM BTA-DNA, 1.5 μM BTA-3OH), 20 nM RE and RI, 1 nM β-lactamase and 10 nM BLIP. c Enzyme activity as a function of time after mixing pre-assembled polymers containing either enzyme and RE or inhibitor and RI. Experiments were performed using final concentrations of 25% BTA-DNA (0.5 μM BTA-DNA, 1.5 μM BTA-3OH), 20 nM RE and RI, 1 nM β-lactamase and 10 nM BLIP. Enzyme activities were normalized to a control omitting the inhibitor protein (green diamonds). Error bars represent the s.d. calculated from triplicate measurements