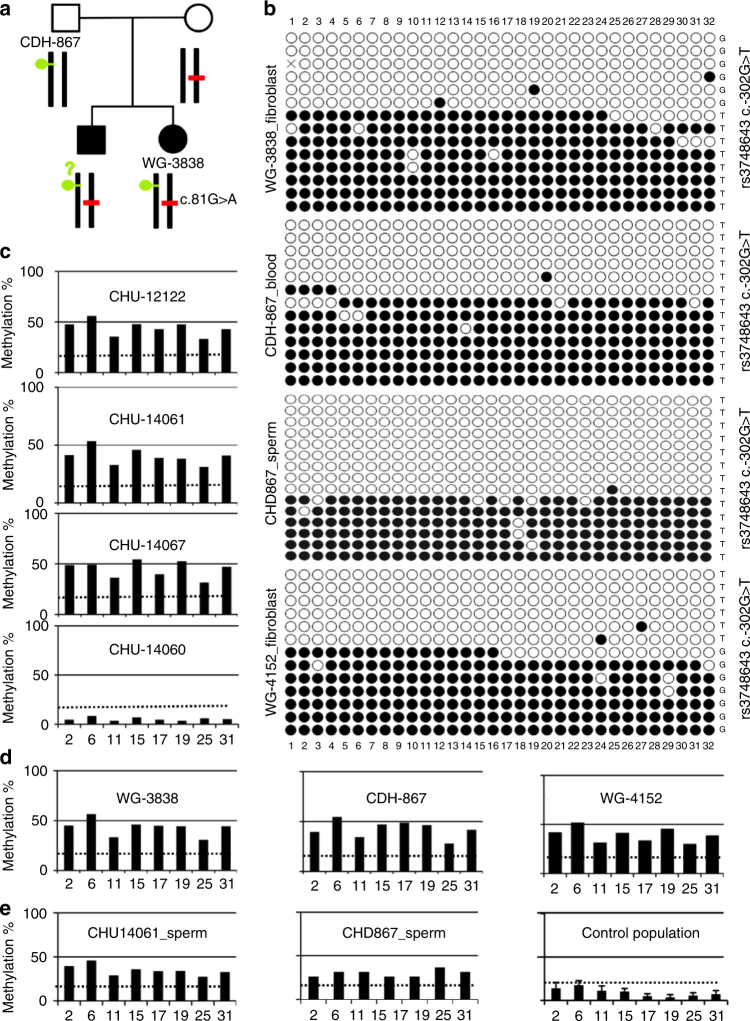
Fig. 2.
The MMACHC epimutation is found in a second case and is present in sperm of both cases. a Pedigree of the family of case WG3838; red bar, mutation (heterozygous c.81G>A splice variant) found in the proband, her mother, and a deceased brother; green circle, epimutation encompassing the MMACHC promotor/exon 1 found in the proband and her mother. No material from the deceased brother was available to study the presence of the epimutation. b MMACHC methylation epigrams after PCR amplification/cloning/Sanger sequencing of the bisulphite-treated DNA from blood obtained from WG-3838, her father CHD867, WG-4152, and from sperm obtained from CHD867. The epimutation was identified in all samples. c HM450K array methylome profiling of the epimutation in the CHU-12122 case and her relatives. The data confirmed the results obtained in Fig. 1c. d Methylome profiling confirms the epimutation in blood DNA from case WG-3838, her father CDH-867, and case WG-4152; the dotted line corresponds to a β value threshold of 0.2, below which the CpG probe was considered fully unmethylated. e Methylome profiling of sperm DNA from the father of case CHU-12122, the father of case WG-3838, and a control population of Utah. The data confirm the epimutation’s presence in the sperm; the dotted line corresponds to a β value threshold of 0.2, below which the CpG probe was considered fully unmethylated. The absence of sperm DNA contamination by DNA from somatic cells was proven by methylation analysis of SNRPN imprinted gene (see Supplementary Fig. 1)