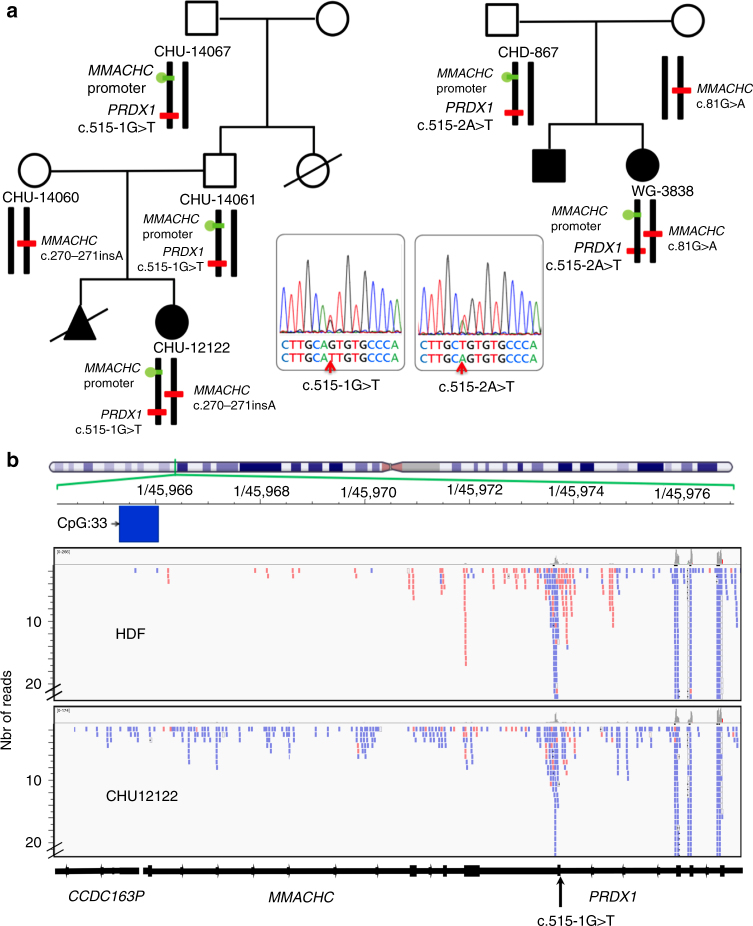
Fig. 5.
Pedigrees derived from whole-genome sequencing and epigenome analyses and RNA-Seq in case fibroblasts. a Pedigrees derived from WGS of DNA by HiSeq X Ten Illumina System in cases CHU-12122 and WG-3838 and their relatives. WGS evidences c.515-1G>T and c.515-2A>T mutations of PRDX1, in cases CHU-12122 and WG-3838 and the relatives who bear the secondary epimutation, respectively. The mutations are absent in relatives, who bear the MMACHC heterozygous mutation. b NGS sequencing of RNA from case CHU-12122 and HDF control fibroblast line. Overlapping antisense (in blue) transcription of PRDX1/MMACHC/CCDC163P trio of genes is predominant in CHU-12122 fibroblasts while MMACHC sense transcription (in red) is predominant in HDF control fibroblasts