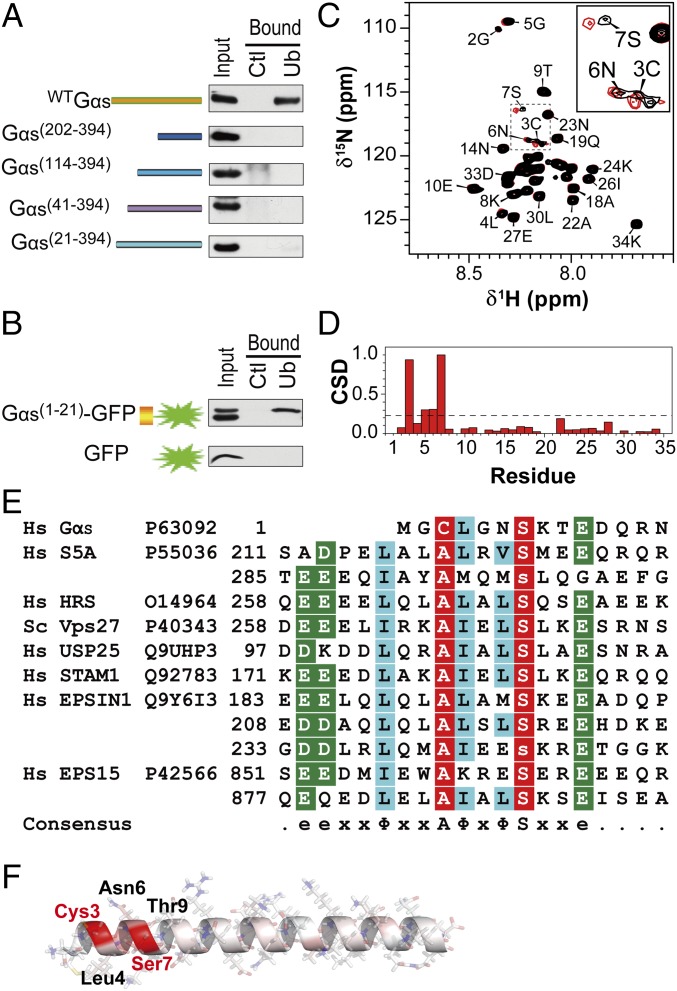
Fig. 2.
Gαs contains a UIM. (A) Mapping the Gαs domain responsible for binding to ubiquitin. HEK293 cell extracts expressing various Gαs N-terminal–truncated mutants were incubated with ubiquitin-agarose beads (Ub) or control protein G-agarose beads (Ctl). (B) Gαs N-terminal residues 1–21 are sufficient for ubiquitin binding. Pull-downs were performed on HEK293 cells expressing GFP alone or fused to the 1–21 region of Gαs. (C) NMR chemical shift mapping of the residues of a Gαs1-34 peptide interacting with ubiquitin. The 1H-15N-HSQC spectrum of Gαs1-34 in the absence (black) and presence (red) of ubiquitin at a 1:10 ratio is shown. Fully assigned spectra are shown in Fig. S3A. (D) Normalized weighted CSDs observed in Gαs1-34 upon ubiquitin binding. Significant displacements are assigned to cross-peaks that have CSD values of 1 SD above the mean (dashed line). (E) Alignment of the 1–14 region of Gαs with the sequences of the UIMs found in various ubiquitin-binding proteins (also Fig. S4). e, acidic residue; x, any amino acid; Φ, large hydrophobic residue. Nearly invariant Ala and Ser positions are colored red, and highly similar amino acids are colored blue and green. Hs, Homo sapiens. (F) Structure of Gαs1-34 peptide modeled by Swiss-PdbViewer and SWISS-MODEL, showing in red the residues with the greatest changes in chemical shift upon addition of ubiquitin.