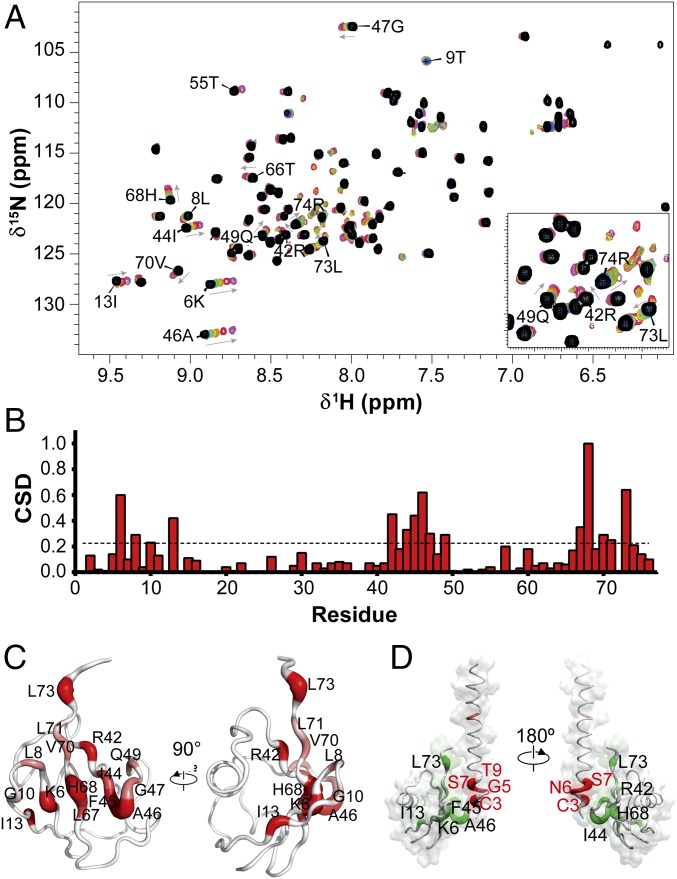
Fig. 3.
Ubiquitin-binding surface of Gαs. (A) NMR chemical shift mapping of the residues in ubiquitin interacting with the Gαs1-34 peptide. The 1H-15N-HSQC spectra of ubiquitin in the absence (black) and increasing ratios (1:1, 1:2.5, 1:5, 1:10, 1:25, 1:50, and 1:100) of Gαs peptide (blue to purple) are shown. Fully assigned spectra are shown in Fig. S3B. (B) Normalized weighted CSD of ubiquitin residues observed upon binding to Gαs1-34 peptide. (C) Surface/worm representation of ubiquitin showing in red the residues with the greatest CSD upon addition of Gαs1-34 peptide. Worm radii are proportional to CSD values, and regions of higher perturbations (red) have a thicker backbone (also Fig. S5). (D) Interaction model mapping the interaction residues on ubiquitin structure and Gαs1-34 peptide. Worm radii are proportional to CSD values and are coded in a white-to-red (Gαs peptide) or white-to-green (ubiquitin) gradient.