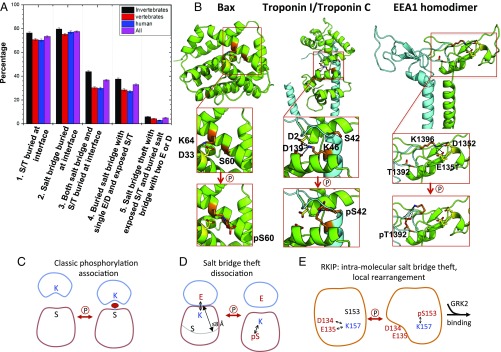
Fig. 5.
Salt-bridge theft mechanism observed in RKIP and other proteins. (A) Bar plot of the frequencies of different interfacial properties among hetero-oligomers on a per-complex basis. Errors are the SD calculated assuming a binomial distribution. See SI Appendix, Table S2, for details. (B) Phosphorylation of Bax at S60 by PKA attracts K64, destabilizing the α-helix and activating translocation and cytochrome c release. Residues involved in the salt bridge (K64, D33) are indicated. Phosphorylation at S42 by PKC prevents the interaction of Troponin I (blue) with Troponin C (green) and inhibits Troponin I activity. Residues involved in the salt bridge are indicated (K46 on chain C; D2 and D139 on chain A). Phosphorylation of EEA1 at T1392 on chain B by the kinase p38 attracts K1396 on chain B, thus freeing D1352 on chain A to interact with phosphatidylinositol-3-phosphate within the endosomal membrane. Residues involved in the salt bridge are indicated (K1396 on chain B; D1352 and possibly E1351 on chain A). (C–E) Alternative models for phosphorylation-controlled protein association.