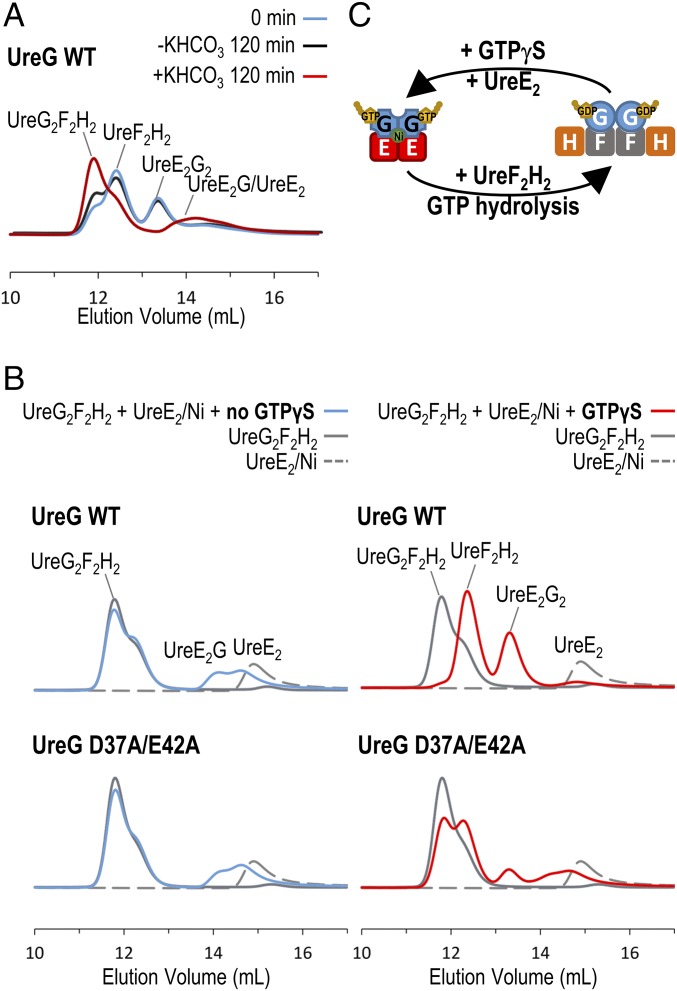
Fig. 4.
UreG swaps protein-binding partners during the GTP hydrolysis/binding cycle. (A) Equal molar ratio (15 µM) mixture of H. pylori UreE2G2/Ni and UreF2H2 complexes (cyan) was incubated in 2 mM MgSO4, 1 mM GTP, 0.2 mM TCEP, 100 mM NaCl, 20 mM Hepes, pH 7.2 with (red) or without (black) 10 mM KHCO3 at 37 °C for 120 min. The protein samples were then analyzed by SEC/SLS. Upon activation of GTP hydrolysis by KHCO3 (red), the UreE2G2 complex disappeared and the majority of the UreF2H2 complex was converted to the UreG2F2H2 complex. (B) A total of 15 µM UreG2F2H2 complex (gray solid lines) and 15 µM UreE2/Ni dimer (gray dotted lines) was added to 2 mM MgSO4, 0.2 mM TCEP, 100 mM NaCl, 20 mM Hepes, pH 7.2 buffer with (red lines) or without (cyan lines) 300 µM GTPγS. The protein samples were analyzed by SEC/SLS. In the absence of GTPγS (Left), only a small amount of UreG dissociated from the UreG2F2H2 complex to form the UreE2G complex. In the presence of GTPγS (Right), wild-type UreG completely dissociated from the UreG2F2H2 complex to form the UreE2G2 complex with the UreE dimer. For the UreG D37A/E42A mutant, the GTP-dependent swapping of protein-binding partners was greatly abolished. (C) Our results suggest that the preference of protein-binding partners is dictated by conformational changes in UreG induced by GTP binding/hydrolysis.