Abstract
MicroRNAs (miRs) are a group of short, endogenous, non-protein-coding and single-stranded RNAs that regulate gene expression by binding to the 3′-untranslated region (3′UTR) of mRNAs, which results in their degradation or translational repression. The aim of the present study was to investigate the expression and function of miR-335 in human papillary thyroid cancer (PTC). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to quantify the relative miR-335 expression levels in PTC tissues and cell lines. The effect of miR-335 on the proliferation, migration and invasion of PTC cells was assessed by an MTT assay, and transwell migration and invasion assays, respectively. Dual-luciferase reporter assays were employed to explore whether miR-335 directly targeted the 3′UTR of the potential target gene zinc finger E-box binding homeobox 2 (ZEB2). RT-qPCR and western blotting were adopted to assess the effect of miR-335 on the mRNA and protein expression of ZEB2. RT-qPCR revealed that miR-335 was downregulated in PTC tissues and cell lines. The MTT assay and transwell migration and invasion assays demonstrated that the overexpression of miR-335 significantly inhibited the proliferation, migration and invasion of PTC cells. ZEB2 was identified as a direct target of miR-335 with computational analysis, which was confirmed with a dual-luciferase reporter assay, RT-qPCR and western blotting. The knockdown of ZEB2 significantly inhibited the proliferation, migration and invasion of PTC cells, indicating that ZEB2 may be a functional target of miR-335. Taken together, these findings suggested that miR-335 functioned as a tumor suppressor and suppressed the growth and metastatic behavior of PTC cells by targeting ZEB2.
Keywords: thyroid cancer, papillary thyroid cancer, microRNA-335, zinc finger E-box binding homeobox 2, growth, metastasis
Introduction
Thyroid cancer is the most common subtype of endocrine malignancy, with 300,000 new cases per year, and ~40,000 mortalities per year, worldwide (1). The incidence of thyroid cancer has increased in recent decades (2). It can be categorized into four major histologic groups, including papillary thyroid cancer (PTC), follicular thyroid cancer, poorly differentiated carcinoma and anaplastic thyroid cancer (3). PTC accounts for 80–90% of all thyroid cancer cases (4).
A number of factors have been demonstrated to be associated with PTC progression, including genetic factors, environmental exposure, epigenetic alteration, nodular disease of the thyroid and radiation exposure (5,6). The prognosis of PTC patients is associated with their age, tumor size, lymph node invasion and distant metastasis (7). Currently, the standard therapeutic treatment for PTC is complete thyroidectomy followed by radioiodine and levothyroxine therapy (8). However, ~10% of patients with PTC develop recurrence and metastasis, which are associated with poor prognosis (9). Thus, it is necessary to characterize the molecular mechanisms of PTC initiation and development, and to develop novel, targeted therapies for PTC.
The abnormal expression of microRNAs (miRNAs/miRs) has been implicated in the pathogenesis of numerous tumor types, including PTC (10–12). miRNAs are a group of short, endogenous, non-protein-coding and single-stranded RNA molecules 18–25 nucleotides in length (13). They function as negative regulators for target mRNA expression through binding to the 3′-untranslated region (3′UTR) of mRNAs in a base-pairing manner, resulting in mRNA degradation or translational repression (14–17). miRNAs have been demonstrated to serve a crucial function in various biological processes, including cell growth, cell cycle, development, differentiation, metabolism and metastasis (18–21). The expression of specific miRNAs may be upregulated in particular types of cancer and downregulated in others; this conflicting observation is predominantly a result of differences in the mechanisms of oncogenesis between tumor types (22,23). Upregulated miRNAs in cancer act as oncogenes by negatively regulating tumor suppressor genes, whereas downregulated miRNAs act as tumor suppressors via the blockade of oncogenes in tumor progression (24,25). Therefore, identifying the targets of miRNAs is essential for understanding the functions of miRNAs in cancer.
In the present study, it was demonstrated that miR-335 was significantly downregulated in PTC tissue and cell lines. In addition, it was identified that miR-335 inhibited PTC cell growth, migration and invasion in vitro. miR-335 may act as a tumor suppressor in PTC by targeting zinc finger E-box binding homeobox 2 (ZEB2). Taken together, the present study revealed a novel perspective on how miR-335 affects PTC.
Materials and methods
Human tissue specimens and ethics
The present study was approved by the research ethics committee of Henan Provincial People's Hospital (Zhengzhou, China). Written informed consent was obtained from the patients with PTC recruited into the study. A total of 59 pairs of human PTC tissues and adjacent normal tissues (NATs) were obtained from Henan Provincial People's Hospital between June 2013 to February 2015. All patients (21 male and 38 female; age range, 32–77 years; mean age, 53 years) enrolled in the study had not received any preoperative treatments, including radiotherapy, chemotherapy and levothyroxine, prior to the thyroidectomy. The tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until use.
Cell culture
The human PTC cell lines (TPC-1, HTH83, K1 and BCPAP) and normal human thyroid cell line (HT-ori3) were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). Although K1 cells are contaminated with GLAG-66 (26), the resulting phenotypic and genotypic differences between K1 and GLAG-66 cells were considered unlikely to affect the outcome of the present study. 293T cells, used for the luciferase reporter assay, were obtained from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All cell lines were maintained in RPMI-1640 or Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 µg/ml streptomycin (all Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), in a humidified incubator with 5% CO2 at 37°C.
miRNA/siRNA transfection
Chemically synthesized miRNA mimics and siRNA [miR-335 mimic, negative control (NC), ZEB2 siRNA and siRNA-ctrl] were acquired from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The miR-335 mimics sequence was 5′-UCAAGAGCAAUAACGAAAAAUGU-3′ and the NC sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. The ZEB2 siRNA sequence was 5′-GGACACAGGUUCUGAAACAdTd T-3′ and the siRNA-ctrl sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. miRNA and siRNA transfection were performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cell lines using the mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For miR-335 expression, the total RNA was reverse transcribed into cDNA by using a TaqMan® MicroRNA Reverse Transcription kit (cat. no., 4366596; Applied Biosystems; Thermo Fisher Scientific, Inc.). A TaqMan miRNA assay (cat. no., 4324018; Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for the quantification of miR-335; U6 was used as an internal control for miR-335 expression. To determine ZEB2 mRNA expression, an M-MLV Reverse Transcription system (Promega Corporation, Madison, WI, USA) was used for reverse transcription according to the manufacturer's protocol, followed by qPCR with SYBR Green Master mix (Takara, Biotechnology Co., Ltd., Dalian, China). The thermocycling conditions for qPCR were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The primers were designed as follows: miR-335, 5′-AGCCGTCAAGAGCAATAACGAA-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); ZEB2, 5′-AGTCCTCCCCACACGTGAGCC-3′ (forward) and 5′-TGCGGTCTGGATCGTGGCTTC-3′ (reverse); and GAPDH, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse). GADPH was used as an internal control for ZEB2 mRNA expression. Each sample was analyzed in triplicate. The relative expression of miRNA and mRNA was analyzed with the 2−∆∆Ct method (27).
Cell viability assay
Cell viability was evaluated with an MTT assay. Cells were seeded into 96-well plates at a density of 3,000 cells per well. Subsequent to incubation overnight, cells were transfected with miRNA or siRNA. At a range of time points subsequent to transfection (24, 48, 72 and 96 h), MTT assays were performed. In brief, 20 µl 5 mg/ml MTT solution (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to each well. The plates were incubated at 37°C for an additional 4 h. Subsequently, the medium was removed and 100 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added to each well to dissolve the purple crystal. The absorbance at 490 nm was detected using an ELISA plate reader. Each sample was analyzed in triplicate.
Transwell migration and invasion assay
The ability of cells to migrate and invade was evaluated with transwell chambers (Corning Incorporated, Corning, NY, USA) with an 8-µm pore polycarbonate membrane insert. miRNA or siRNA-transfected cells were treated with trypsin/EDTA solution, washed once with serum-free culture medium and re-suspended in serum-free culture medium. 5×104 cells in 200 µl serum-free medium were seeded into the upper chamber, and 500 µl medium containing 20% FBS was added into the lower chamber. A cell invasion assay was performed with the same procedure, with the exception that the transwell chamber membranes were pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA). Subsequent to incubation at 37°C for 48 h, cells that migrated or invaded to the bottom surface of the transwell chambers were fixed with 100% methanol at room temperature for 10 min, stained with 0.5% crystal violet at room temperature for 10 min and washed with PBS three times. Cells on the top surface of the transwell chamber were removed with a cotton swab. The migration and invasion abilities were quantified by counting the number of migrated and invaded cells in five fields per transwell chamber, using an inverted microscope (×200 magnification; Olympus Corporation, Tokyo, Japan). Each experiment was repeated three times.
Western blotting
Total protein was extracted from cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) containing protease inhibitors. Total protein concentration was detected with a Bicinchoninic Acid Protein assay kit (Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. Equivalent amounts of protein (20 µg per lane) were separated via 10% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA) using a semidry transfer system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Subsequent to blocking with 5% skimmed milk in Tris buffered saline with Tween-20 (TBST) at room temperature for 1 h, the membranes were probed with the primary antibodies, including monoclonal mouse anti-human ZEB2 (dilution, 1:500; cat. no., sc-271984) and β-actin (dilution, 1:500; cat. no., sc-47778; both Santa Cruz Biotechnology, Dallas, TX, USA) antibodies, overnight at 4°C. Subsequent to washing with TBST three times, the membranes were incubated with a goat anti-mouse horseradish peroxidase-conjugated secondary antibody (dilution, 1:5,000; cat. no., sc-2005; Santa Cruz Biotechnology) at room temperature for 2 h, followed by development with ECL Plus reagent (Pierce; Thermo Fisher Scientific, Inc.). β-actin was used as an internal control. This assay was repeated three times.
Dual-luciferase reporter assay
TargetScan (http://www.targetscan.org/) was used to predict targets of miR-335. Luciferase reporter plasmids, pGL3-ZEB2-3′UTR wild type (Wt) and pGL3-ZEB2-3′UTR mutant (Mut), were synthesised by Shanghai GenePharma Co., Ltd (Shanghai, China). For the luciferase reporter assay, 293T cells were seeded into 24-well plates at a density of 1.5×105 cells per well. Following incubation overnight, cells were co-transfected with pGL3-ZEB2-3′UTR Wt or pGL3-ZEB2-3′UTR, and miR-335 or NC, using Lipofectamine 2000. At 48 h post-transfection, firefly and Renilla luciferase activity were measured using a Dual-Luciferase Reporter assay system (Promega Corporation). The Renilla luciferase activity was measured as an internal control for each well. The assay was performed in triplicate.
Statistical analysis
Data were expressed as the mean ± standard deviation, and compared with a Student's t-test or one-way analysis of variance using SPSS 17 software (SPSS Inc., Chicago, IL, USA). Student-Newman-Keuls test was used as a post hoc test following ANOVA. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-335 is downregulated in PTC tissues and cell lines
In order to confirm the association of miR-335 with PTC, miR-335 expression levels were investigated in PTC tissue and matched NATs. RT-qPCR analysis revealed that miR-335 was significantly downregulated in PTC tissue compared with the matched NATs (Fig. 1A; P<0.05).
Figure 1.
miR-335 is downregulated in PTC tissues and cell lines. (A) miR-335 levels were relatively low in PTC tissues compared with adjacent tissue samples, based on reverse transcription-quantitative polymerase chain reaction data. (B) miR-335 was downregulated in PTC cell lines in comparison to normal human thyroid HT-ori3 cells. *P<0.05 compared with the respective controls. miR, microRNA; PTC, papillary thyroid cancer.
In addition, the expression levels of miR-335 in four PTC cell lines and a normal human thyroid cell line, HT-ori3, were also quantified. The results indicated that the miR-335 expression levels were reduced in the four PTC cell lines relative to the expression in HT-ori3 (Fig. 1B; P<0.05). These results suggested that miR-335 may serve a role in the development of PTC.
miR-335 is upregulated in HTH83 and K1 cells following transfection with an miR-335 mimic
miR-335 expression was the lowest in HTH83 and K1 cells. Therefore, HTT83 and K1 cells were selected for functional studies. To explore the function of miR-335 in PTC, an miR-335 mimic or NC was transfected into HTT83 and K1 cells. The transfection efficiency was assessed with RT-qPCR. The result indicated that miR-335 was significantly upregulated in HTH83 and K1 cells following transfection with an miR-335 mimic compared with transfection with the NC (Fig. 2; P<0.05).
Figure 2.

miR-335 is upregulated in HTH83 and K1 cells following transfection with an miR-335 mimic. *P<0.05 compared with NC. miR, microRNA; NC, negative control.
Overexpression of miR-335 suppresses the proliferation, migration and invasion of PTC cells
An MTT assay was performed to evaluate the effect of miR-335 on PTC cell proliferation. The induced overexpression of miR-335 in HTH83 and K1 cells resulted in a reduced proliferation rate compared with the NC groups (Fig. 3A; P<0.05).
Figure 3.
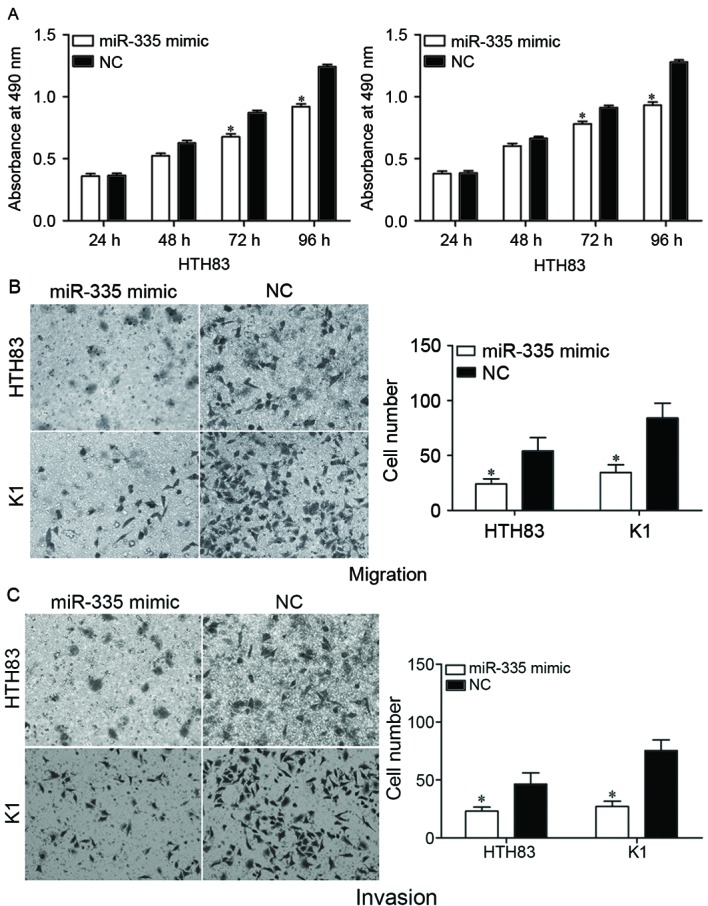
miR-335 overexpression inhibits the proliferation, migration and invasion of HTH83 and K1 cells. The upregulation of miR-335 significantly inhibited the (A) proliferation, (B) migration and (C) invasion of HTH83 and K1 cells, as determined by an MTT assay and transwell migration and invasion assays, respectively *P<0.05 compared with NC. miR, microRNA; NC, negative control.
The effect of miR-335 on cell migration and invasion was assessed by using transwell migration and invasion assays. The results indicated that the upregulation of miR-335 decreased the migration (Fig. 3B; P<0.05) and invasion (Fig. 3C; P<0.05) abilities of HTH83 and K1 cells. Collectively, the data suggested that miR-335 was a regulator of proliferation, migration and invasion in PTC cells.
ZEB2 is a direct target of miR-335
TargetScan was used to predict the direct target genes of miR-335. As illustrated in Fig. 4A, the ZEB2 mRNA 3′UTR contained a predicted binding site for miR-335. To explore whether miR-335 directly targeted the 3′UTR of ZEB2, a dual-luciferase reporter assay was performed. The luciferase activity was significantly reduced in 293T cells co-transfected with ZEB2-3′UTR Wt and the miR-335 mimic compared with the cells co-transfected with ZEB2-3′UTR Mut and the miR-335 mimic (Fig. 4B; P<0.05).
Figure 4.
ZEB2 is a direct target of miR-335 in PTC. (A) The miR-335 binding site in the 3′UTR of ZEB2, and the ZEB2-3′UTR Mut sequence. (B) Luciferase activity was detected in 293T cells following co-transfection with the ZEB2-3′UTR Wt or ZEB2-3′UTR Mut with an miR-335 mimic or NC. (C) Reverse transcription-quantitative polymerase chain reaction was performed to determine ZEB2 mRNA expression levels in HTH83 and K1 cells. (D) Western blotting was performed to assess ZEB2 protein expression levels in HTH83 and K1 cells. *P<0.05 compared with NC. ZEB2, zinc finger E-box binding homeobox 2; miR, microRNA; PTC, papillary thyroid cancer; 3′UTR, 3′-untranslated region; Mut, mutant; Wt, wild-type; NC, negative control.
To determine whether there was an effect of miR-335 expression on ZEB2 expression, HTH83 and K1 cells transfected with miR-335 mimics were evaluated with RT-qPCR and western blot. RT-qPCR analysis indicated that the overexpression of miR-335 reduced ZEB2 mRNA levels in HTH83 and K1 cells (Fig. 4C; P<0.05). The western blot analysis revealed that the expression of ZEB2 protein was downregulated in miR-335 mimic-transfected HTH83 and K1 cells (Fig. 4D; P<0.05). Taken together, the results demonstrated that ZEB2 is a direct target of miR-335 in PTC.
miR-335 inhibits the proliferation, migration and invasion of PTC cells via the regulation of ZEB2
ZEB2 was demonstrated to be a direct target of miR-335 in PTC. Therefore, we hypothesized that miR-335 may have decreased the proliferation, migration and invasion of HTH83 and K1 cells by the downregulation of ZEB2. To confirm this, ZEB2 siRNA was used to knockdown ZEB2 expression. Following transfection, western blot analysis demonstrated that ZEB2 was significantly downregulated in HTH83 and K1 cells (Fig. 5A; P<0.05). The effect of ZEB2 siRNA on the proliferation, migration and invasion of PTC cells was also measured. The results indicated that ZEB2 siRNA significantly suppressed cellular proliferation (Fig. 5B; P<0.05), migration (Fig. 5C; P<0.05) and invasion (Fig. 5D; P<0.05) compared with negative control siRNA groups. These data suggested that the overexpression of miR-335 inhibited the proliferation, migration and invasion of PTC cells at least partially by the knockdown of ZEB2 expression.
Figure 5.
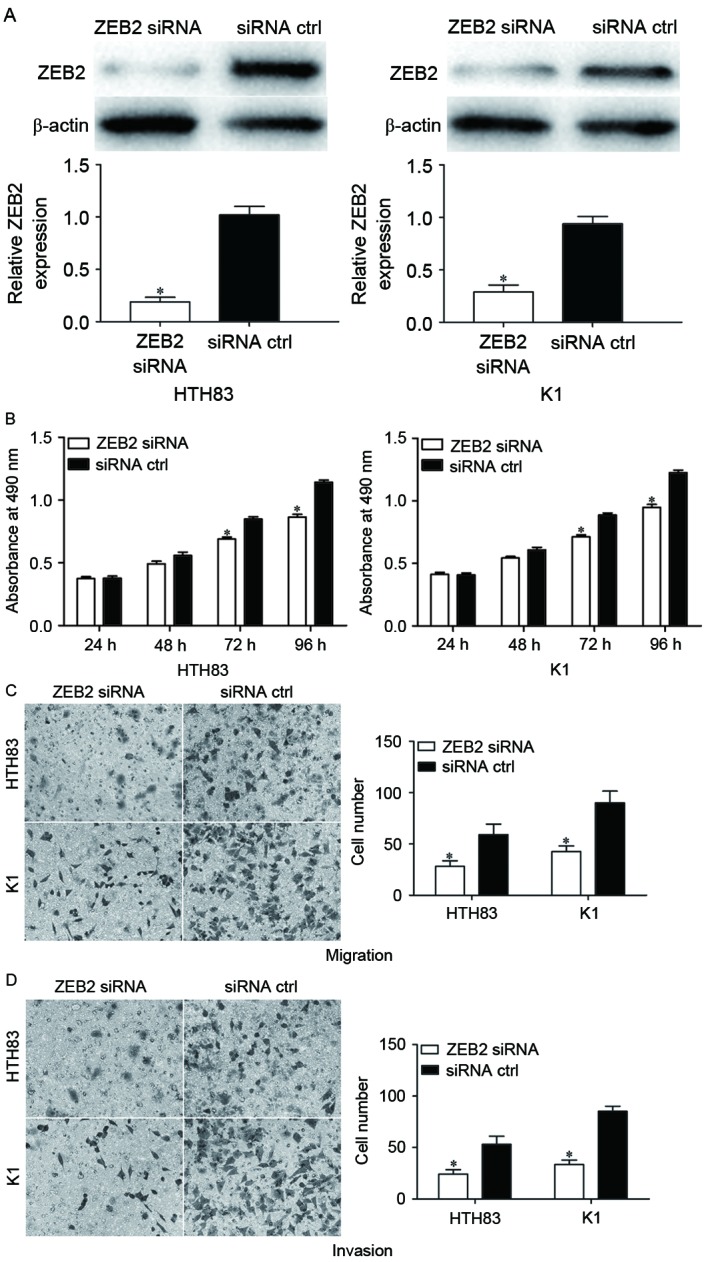
Knockdown of ZEB2 suppresses the proliferation, migration and invasion of HTH83 and K1 cells. (A) ZEB2 siRNA was demonstrated to reduce the ZEB2 expression levels in HTH83 and K1 cells. (B) It was demonstrated with an MTT assay that ZEB2 siRNA suppressed the proliferation of HTH83 and K1 cells. Transwell (C) migration and (D) invasion assays indicated that ZEB2 siRNA significantly suppressed the migration and invasion of HTH83 and K1 cells. *P<0.05 compared with siRNA ctrl. ZEB2, zinc finger E-box binding homeobox 2; ZEB2 siRNA, small interfering RNA against ZEB2; siRNA ctrl, small interfering RNA control.
Discussion
It has previously been demonstrated that alterations in the expression of miRNAs occur in a large variety of tumor types in humans (28). There may be a connection between the functions of miRNAs and carcinogenesis, which is supported by investigations of the expression and functions of miRNAs in cancer tissue specimens (29). In the present study, it was identified that that miR-335 was significantly downregulated in PTC tissues and cell lines compared with matched NATs and normal thyroid cells, respectively. Functional studies revealed that miR-335 may have functioned as a tumor suppressor in PTC cells, as its expression was associated with the inhibition of cell growth, migration and invasion. In addition, ZEB2 was identified as a direct target gene of miR-335 in PTC. These results suggest that miRNA-335 may be a candidate in the therapy of patients with PTC.
miR-335, located at 7q32.2, has also been demonstrated as a tumor suppressor in other types of cancer. For example, the overexpression of miR-335 was observed to suppress breast cancer cell proliferation, cell cycle progress, colony formation and cell invasion through the negative regulation of Paired box 6 (30). Xu et al (31) reported that the expression level of miR-335 was relatively low in gastric cancer tissues, and that a low miR-335 expression level was correlated with lymph node metastasis, an advanced pT and pN stage, and the invasion of lymphatic vessels. The upregulation of miR-335 may have inhibited gastric cancer cell invasion and metastasis in vitro and in vivo by directly targeting SP1, as well as acting indirectly through the Bcl-W-induced phosphoinositide 3-kinase-Akt-SP1 pathway (31). The reduced expression of miR-335 was also identified in human prostate cancer tissues and cell lines; miR-335 expression level was associated with a high Gleason Score, advanced clinical stage and metastasis in patients with prostate cancer (32). Enforced miR-335 expression decreased the growth and metastatic characteristics of prostate cancer cells in vitro (32). Sun et al (33) identified that miR-335 was downregulated in more invasive colorectal cancer tissues and cell lines. Kaplan-Meier survival analysis suggested that colorectal cancer tumors with a low miR-335 expression level were associated with a reduced overall survival time. Furthermore, the upregulation of miR-335 suppressed cancer cell migration and invasion in vitro and metastasis to the lung and liver in vivo via the inhibition of ZEB2 (33). These findings indicated that miR-335 may serve as a potential therapeutic target in the treatment of these cancers.
Studies have also indicated that miR-335 is upregulated in meningioma (34) and glioma (35) tumors. The ectopic expression of miR-335 promoted cell growth and prevented cell cycle arrest in the G0/G1 phase through directly targeting the Rb1 signaling pathway in meningioma (34). Jiang et al (35) identified that a relatively high miR-335 expression level was associated with advanced tumor progression in glioma. Furthermore, miR-335 expression was verified for the first time as an independent prognostic marker for predicting the clinical outcome of patients with glioma (35). These results appear to be conflicting, as miR-335 was demonstrated as an oncogene in certain types of cancer and a tumor suppressor in others. This contradiction may be explained by the ‘imperfect complementarity’ of the interactions between miRNAs and target mRNAs (36).
The identification of cancer-specific miRNAs and their target genes may provide therapeutic targets for PTC. To investigate the molecular mechanism of miR-335 in PTC, bioinformatics analysis was performed to predict potential target genes. ZEB2 was predicted as a target gene for miR-335. To determine whether miR-335 directly targets ZEB2, a dual-luciferase reporter assay was performed; it was revealed that miR-335 significantly decreased the luciferase activity of cells transfected with ZEB2-3′UTR Wt compared with cells transfected with ZEB2-3′UTR Mut. The upregulation of miR-335 suppressed the mRNA and protein expression levels of ZEB2 in PTC cells. The effect of ZEB2 siRNA in PTC cells was similar to the effect of miR-335, which suggested that ZEB2 may be a functional target of miR-335 in PTC.
ZEB2, a member of the zinc finger family, functions as a transcriptional repressor of E-cadherin (37). ZEB2 has been identified as upregulated in various types of human cancer, including breast, gastric, head and neck, hepatocellular, ovarian and non-small cell lung carcinoma, and glioma (38–44). ZEB2 was previously identified as upregulated in PTC cell lines compared with normal thyroid cancer cells (45). ZEB2 was also previously identified as contributing to thyroid cancer migration and invasion (3). In the present study, it was demonstrated that the downregulation of ZEB2 significantly inhibited the growth, migration and invasion of PTC cells. These results indicate that ZEB2 may require scrutiny as a potential target for the inhibition of PTC growth and metastasis.
In conclusion, miR-335 was downregulated in PTC tissue samples and cell lines compared with matched NATs and normal thyroid cells, respectively. The overexpression of miR-335 suppressed the proliferation, migration and invasion of PTC cells. Furthermore, ZEB2 may be a functional target of miR-335 in PTC. All the results obtained in the present study suggest that miR-335 may serve as a tumor suppressor gene in the tumorigenesis and progression of PTC. The upregulation of miR-335 may be a novel potential therapeutic strategy in the treatment of patients with PTC.
References
- 1.Sondermann A, Andreghetto FM, Moulatlet AC, da Silva Victor E, de Castro MG, Nunes FD, Brandão LG, Severino P. MiR-9 and miR-21 as prognostic biomarkers for recurrence in papillary thyroid cancer. Clin Exp Metastasis. 2015;32:521–530. doi: 10.1007/s10585-015-9724-3. [DOI] [PubMed] [Google Scholar]
- 2.Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381:1058–1069. doi: 10.1016/S0140-6736(13)60109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan H, Liang W, Xie Z, Li H, Liu J, Liu L, Xiu L, Li Y. Down-regulation of miR-144 promotes thyroid cancer cell invasion by targeting ZEB1 and ZEB2. Endocrine. 2015;48:566–574. doi: 10.1007/s12020-014-0326-7. [DOI] [PubMed] [Google Scholar]
- 4.Geraldo MV, Fuziwara CS, Friguglieti CU, Costa RB, Kulcsar MA, Yamashita AS, Kimura ET. MicroRNAs miR-146-5p and let-7f as prognostic tools for aggressive papillary thyroid carcinoma: A case report. Arq Bras Endocrinol Metabol. 2012;56:552–557. doi: 10.1590/S0004-27302012000800015. [DOI] [PubMed] [Google Scholar]
- 5.Zheng H, Wang M, Jiang L, Chu H, Hu J, Ning J, Li B, Wang D, Xu J. BRAF-activated long noncoding RNA modulates papillary thyroid carcinoma cell proliferation through regulating thyroid stimulating hormone receptor. Cancer Res Treat. 2016;48:698–707. doi: 10.4143/crt.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider AB, Sarne DH. Long-term risks for thyroid cancer and other neoplasms after exposure to radiation. Nat Clin Pract Endocrinol Metab. 2005;1:82–91. doi: 10.1038/ncpendmet0022. [DOI] [PubMed] [Google Scholar]
- 7.Deng X, Wu B, Xiao K, Kang J, Xie J, Zhang X, Fan Y. MiR-146b-5p promotes metastasis and induces epithelial-mesenchymal transition in thyroid cancer by targeting ZNRF3. Cell Physiol Biochem. 2015;35:71–82. doi: 10.1159/000369676. [DOI] [PubMed] [Google Scholar]
- 8.Chou CK, Yang KD, Chou FF, Huang CC, Lan YW, Lee YF, Kang HY, Liu RT. Prognostic implications of miR-146b expression and its functional role in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98:E196–E205. doi: 10.1210/jc.2012-2666. [DOI] [PubMed] [Google Scholar]
- 9.Lang BH, Wong KP, Wan KY, Lo CY. Significance of metastatic lymph node ratio on stimulated thyroglobulin levels in papillary thyroid carcinoma after prophylactic unilateral central neck dissection. Ann Surg Oncol. 2012;19:1257–1263. doi: 10.1245/s10434-012-2385-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi J, Rice SJ, Salzberg AC, Runkle EA, Liao J, Zander DS, Mu D. MiR-365 regulates lung cancer and developmental gene thyroid transcription factor 1. Cell Cycle. 2012;11:177–186. doi: 10.4161/cc.11.1.18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang HG, Luo X, Wu S, Jian B. MiR-99a inhibits cell proliferation and tumorigenesis through targeting mTOR in human anaplastic thyroid cancer. Asian Pac J Cancer Prev. 2015;16:4937–4944. doi: 10.7314/APJCP.2015.16.12.4937. [DOI] [PubMed] [Google Scholar]
- 12.Boufraqech M, Zhang L, Jain M, Patel D, Ellis R, Xiong Y, He M, Nilubol N, Merino MJ, Kebebew E. miR-145 suppresses thyroid cancer growth and metastasis and targets AKT3. Endocr Relat Cancer. 2014;21:517–531. doi: 10.1530/ERC-14-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 14.Nikiforova MN, Gandhi M, Kelly L, Nikiforov YE. MicroRNA dysregulation in human thyroid cells following exposure to ionizing radiation. Thyroid. 2011;21:261–266. doi: 10.1089/thy.2010.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 16.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 17.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 18.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 19.Schmittgen TD. Regulation of microRNA processing in development, differentiation and cancer. J Cell Mol Med. 2008;12:1811–1819. doi: 10.1111/j.1582-4934.2008.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rottiers V, Najafi-Shoushtari SH, Kristo F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N, Mostoslavsky R, Näär AM. MicroRNAs in metabolism and metabolic diseases. Cold Spring Harb Symp Quant Biol. 2011;76:225–233. doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 23.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang T, Thakur A, Chen T, Yang L, Lei G, Liang Y, Zhang S, Ren H, Chen M. MicroRNA-15a induces cell apoptosis and inhibits metastasis by targeting BCL2L2 in non-small cell lung cancer. Tumour Biol. 2015;36:4357–4365. doi: 10.1007/s13277-015-3075-1. [DOI] [PubMed] [Google Scholar]
- 26.Ribeiro FR, Meireles AM, Rocha AS, Teixeira MR. Conventional and molecular cytogenetics of human non-medullary thyroid carcinoma: Characterization of eight cell line models and review of the literature on clinical samples. BMC Cancer. 2008;8:371. doi: 10.1186/1471-2407-8-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Dassow H, Aigner A. MicroRNAs (miRNAs) in colorectal cancer: From aberrant expression towards therapy. Curr Pharm Des. 2013;19:1242–1252. doi: 10.2174/1381612811319070008. [DOI] [PubMed] [Google Scholar]
- 30.Meng Y, Zou Q, Liu T, Cai X, Huang Y, Pan J. microRNA-335 inhibits proliferation, cell-cycle progression, colony formation and invasion via targeting PAX6 in breast cancer cells. Mol Med Rep. 2015;11:379–385. doi: 10.3892/mmr.2014.2684. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang X, Jiang L, Sun Z, Miao Z, Xu H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong SW, Lin TX, Xu KW, Dong W, Ling XH, Jiang FN, Chen G, Zhong WD, Huang J. MicroRNA-335 acts as a candidate tumor suppressor in prostate cancer. Pathol Oncol Res. 2013;19:529–537. doi: 10.1007/s12253-013-9613-5. [DOI] [PubMed] [Google Scholar]
- 33.Sun Z, Zhang Z, Liu Z, Qiu B, Liu K, Dong G. MicroRNA-335 inhibits invasion and metastasis of colorectal cancer by targeting ZEB2. Med Oncol. 2014;31:982. doi: 10.1007/s12032-014-0982-8. [DOI] [PubMed] [Google Scholar]
- 34.Shi L, Jiang D, Sun G, Wan Y, Zhang S, Zeng Y, Pan T, Wang Z. miR-335 promotes cell proliferation by directly targeting Rb1 in meningiomas. J Neurooncol. 2012;110:155–162. doi: 10.1007/s11060-012-0951-z. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J, Sun X, Wang W, Jin X, Bo X, Li Z, Bian A, Jiu J, Wang X, Liu D, et al. Tumor microRNA-335 expression is associated with poor prognosis in human glioma. Med Oncol. 2012;29:3472–3477. doi: 10.1007/s12032-012-0259-z. [DOI] [PubMed] [Google Scholar]
- 36.Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y, Yu W, Wu X, Ye J, Yang S, et al. Identification of miR-7 as an oncogene in renal cell carcinoma. J Mol Histol. 2013;44:669–677. doi: 10.1007/s10735-013-9516-5. [DOI] [PubMed] [Google Scholar]
- 37.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bindels S, Mestdagt M, Vandewalle C, Jacobs N, Volders L, Noël A, van Roy F, Berx G, Foidart JM, Gilles C. Regulation of vimentin by SIP1 in human epithelial breast tumor cells. Oncogene. 2006;25:4975–4985. doi: 10.1038/sj.onc.1209511. [DOI] [PubMed] [Google Scholar]
- 39.Kurashige J, Kamohara H, Watanabe M, Hiyoshi Y, Iwatsuki M, Tanaka Y, Kinoshita K, Saito S, Baba Y, Baba H. MicroRNA-200b regulates cell proliferation, invasion and migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg Oncol. 2012;19(Suppl 3):S656–S664. doi: 10.1245/s10434-012-2217-6. [DOI] [PubMed] [Google Scholar]
- 40.Chu PY, Hu FW, Yu CC, Tsai LL, Yu CH, Wu BC, Chen YW, Huang PI, Lo WL. Epithelial-mesenchymal transition transcription factor ZEB1/ZEB2 co-expression predicts poor prognosis and maintains tumor-initiating properties in head and neck cancer. Oral Oncol. 2013;49:34–41. doi: 10.1016/j.oraloncology.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Qi S, Song Y, Peng Y, Wang H, Long H, Yu X, Li Z, Fang L, Wu A, Luo W, et al. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion and apoptosis in glioma. PLoS One. 2012;7:e38842. doi: 10.1371/journal.pone.0038842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai MY, Luo RZ, Chen JW, Pei XQ, Lu JB, Hou JH, Yun JP. Overexpression of ZEB2 in peritumoral liver tissue correlates with favorable survival after curative resection of hepatocellular carcinoma. PLoS One. 2012;7:e32838. doi: 10.1371/journal.pone.0032838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Q, Guo R, Lin M, Zhou B, Wang Y. MicroRNA-200a inhibits CD133/1+ ovarian cancer stem cells migration and invasion by targeting E-cadherin repressor ZEB2. Gynecol Oncol. 2011;122:149–154. doi: 10.1016/j.ygyno.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 44.Gemmill RM, Roche J, Potiron VA, Nasarre P, Mitas M, Coldren CD, Helfrich BA, Garrett-Mayer E, Bunn PA, Drabkin HA. ZEB1-responsive genes in non-small cell lung cancer. Cancer Lett. 2011;300:66–78. doi: 10.1016/j.canlet.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montemayor-Garcia C, Hardin H, Guo Z, Larrain C, Buehler D, Asioli S, Chen H, Lloyd RV. The role of epithelial mesenchymal transition markers in thyroid carcinoma progression. Endocr Pathol. 2013;24:206–212. doi: 10.1007/s12022-013-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]




