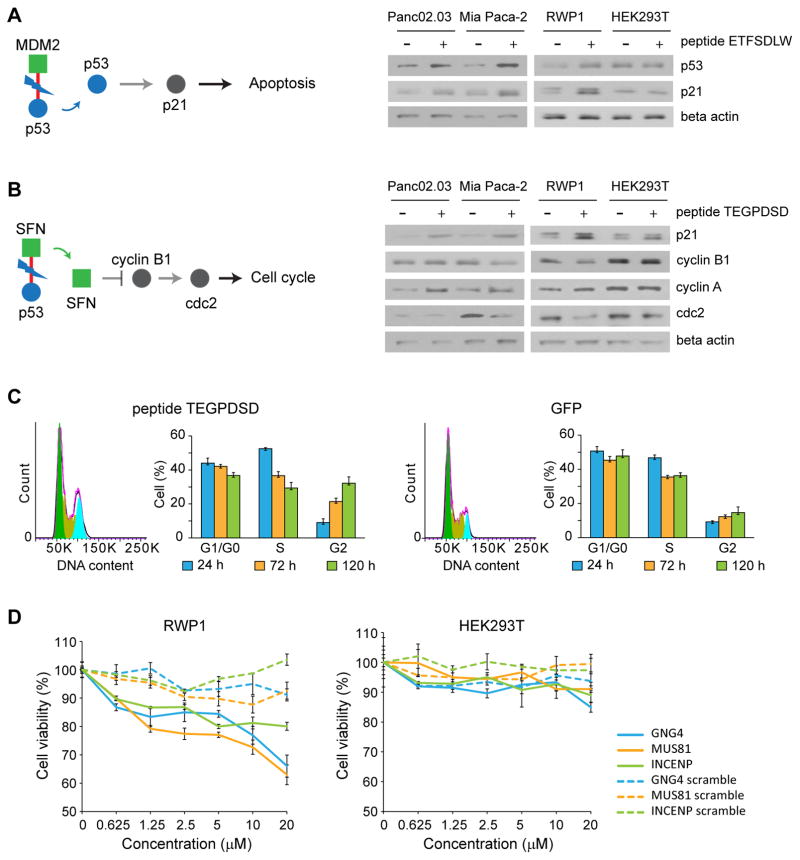
Figure 4. Mechanism of action of cancer-specific inhibitors.
Downstream effect of peptide (A) ETFSDLW (inhibiting P53/MDM2). As is documented, disruption of this interaction leads to a decrease in degradation of P53, thereby increasing P53 levels, triggering apoptosis.. Experiments were done in triplicate. Full gel images in Supplementary Fig. 11. (B) TEGPDSD (inhibiting p53/SFN). Inhibition of this interaction appears to lead to decrease in cyclin B1 and cdc2 levels (and an increase in cyclin A), suggesting it may trigger cell cycle arrest. Blue circle represents source protein of peptide, and green square represents targets of peptide. Red line represents interaction between cancer-specific inhibitor and target. Experiments were done in triplicate. Full gel images in Supplementary Fig. 11. (C) Effect of peptide TEGPDSD on cell cycle. Fluorescence-activated cell sorting (FACS) analysis confirms that it leads to cell cycle arrest in G2. Experiments were done in triplicate. Data represent mean values ± s.d. (D) Effect of synthetic lipidated peptides on cell viability. Synthetic GNG4, MUS81 and INCENP derived lipopeptides show strong dosage dependent effects on cell viability in RWP1 cells, while scrambled versions do not affect cell viability. Experiments were done in duplicate. Data represent mean values ± s.d.