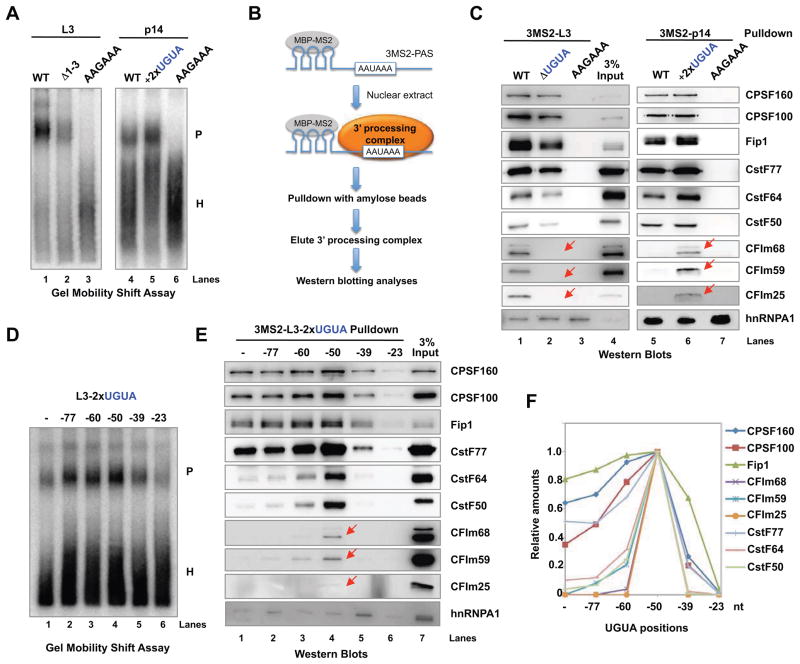
Figure 3. Enhancer-bound CFIm promotes the assembly of mRNA 3′ processing complex.
(A) Gel mobility shift assays with L3 or p14-derived PASs. P: mRNA 3′ processing complex; H: heterogenous complex. (B) A diagram showing the RNA affinity purification procedure. MBP-MS2: a fusion protein between maltose binding protein and MS2. (C) The complexes assembled on the 3MS2-tagged L3 or p14-derived PASs were purified and analyzed by western blotting. The red arrows mark the CFIm subunits. (D) mRNA 3′ processing complex assembly on L3-derived PASs as shown in Fig. 2(A). (E) The mRNA 3′ processing complexes assembled on the 3MS2-tagged L3 derivatives as shown in Fig. 2(A) were purified and analyzed by western blotting. (F) Quantification of western blot signals in (E) using ImageJ.