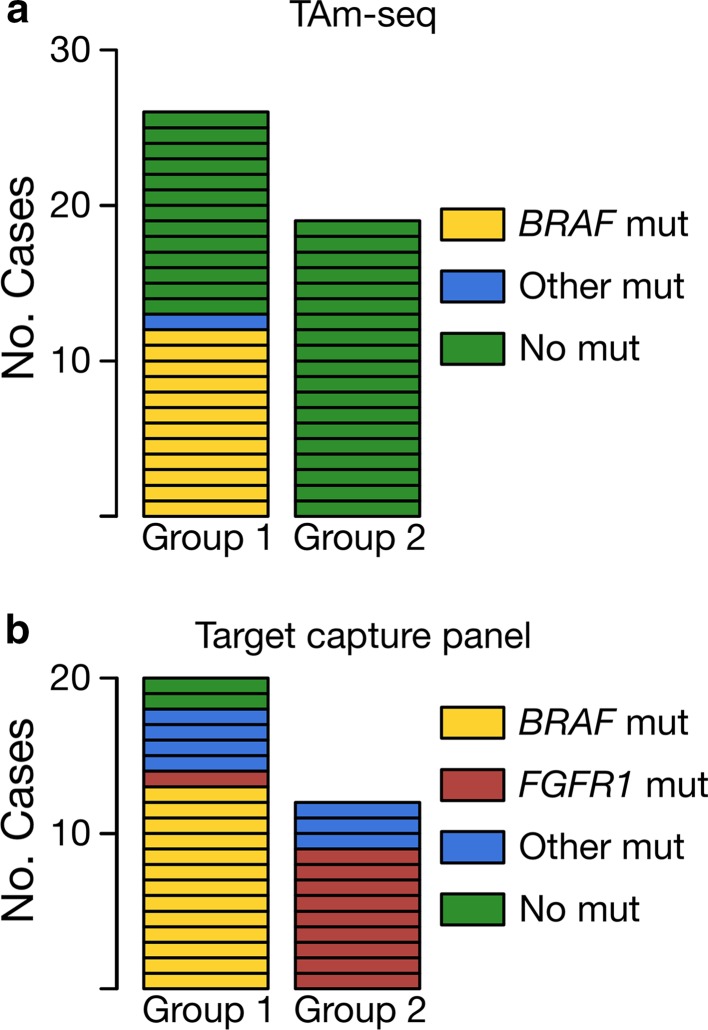
Fig. 3.
Majority of glioneuronal tumours possess pathogenic mutations; BRAF and FGFR1 mutations are highly selective for groups identified by consensus clustering. a Tagged amplicon sequencing against 45 tumours assayed for mutations in CTNNB1, BRAF, HIST1H3B, H3F3A, IDH1, and IDH2. 14/45 (31%) possessed a mutation, of which the most common was BRAF-V600E in 12/26 (46%) Group 1 tumours. No BRAF mutations were detected in Group 2 tumours. Mutations in CTNNB1, H3F3A, and IDH1 were detected in 4 cases, 1 of which was BRAF-V600E positive. b Target capture DNA sequencing against 32 tumours assayed for a panel of 78 genes. 30/32 (94%) possessed mutations. The most common abnormalities were BRAF-V600E, affecting 13/20 (65%) Group 1 tumours, and abnormalities in FGFR1 in 1/20 (5%) Group 1 and 9/12 (75%) Group 2 tumours. Mutations in ATM, TP53, AKT1, MAP2K2, HRAS, APC, WT1, ARID1A, ACVR1, MYCN, and CDK4 were also detected both independently and alongside BRAF and FGFR1 abnormalities but were not recurrent