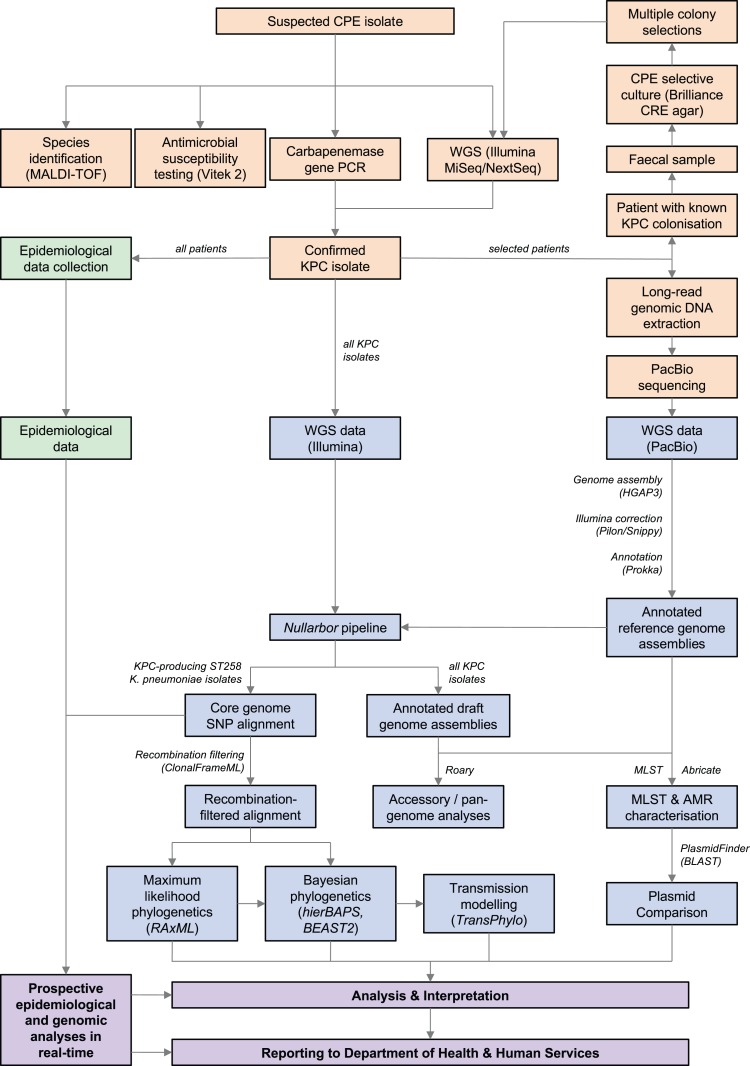
Figure 1. Workflow summary diagram of methods used in this study.
Microbiology methods (orange), bioinformatic methods (blue), and epidemiological methods (green) used to generate data for analysis and reporting to public health authorities (purple) are shown. Results of combined prospective epidemiological and genomic analyses performed iteratively during the outbreak were reported to the Department of Health and Human Services in real-time. CPE, carbapenemase-producing Enterobacteriaceae; KPC, Klebsiella pneumoniae carbapenemase; MALDI-TOF, matrix-assisted laser desorption/ionisation time-of-flight; PCR, polymerase chain reaction; WGS, whole-genome sequencing; SNP, single nucleotide polymorphism; AMR, antimicrobial resistance; MLST, multi-locus sequence typing.