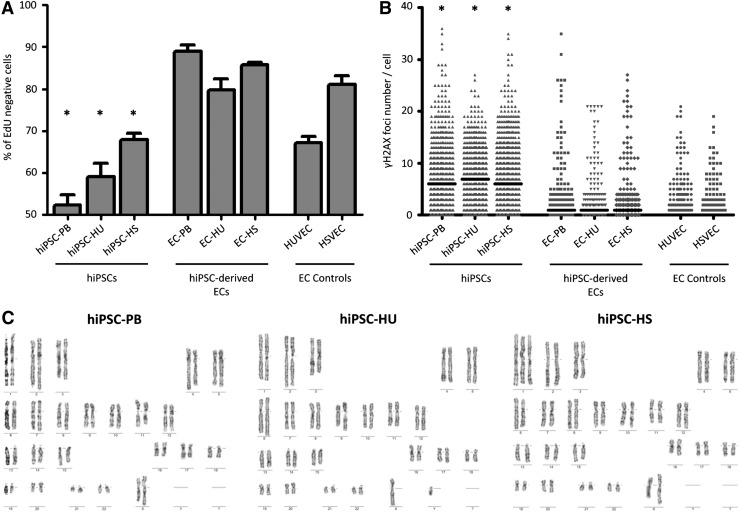
FIG. 5.
Cell cycle speed and genome stability during the reprogramming and differentiation processes. (A) Percentage of EdU-negative (G1 phase) cells among hiPSCs, hiPSC-derived ECs, and EC controls. Mean value (±SEM; n = 3). Asterisks indicate statistically significant (P < 0.05) decreases in the percentages of hiPSCs in G1 phase compared with their EC counterparts, as detected using Student's t-test. (B) Number of γH2AX foci per cell in hiPSCs, hiPSC-derived ECs and EC controls. The bar represents the median. Asterisks indicate statistically significant (P < 0.05) differences between hiPSCs and ECs, as confirmed by the Mann–Whitney test. (C) Cytogenetic data from hiPSC lines. Approximately 100% of cells possess a normal karyotype in hiPSC-PB and hiPSC-HU lines (passages 27 and 16, respectively). Representative aneuploid karyotype detected in 80% of cells in the hiPSC-HS line, in which a gain of chromosome 1 was observed (passage 17). EdU, 5-ethynyl-2′-deoxyuridine; γH2AX, phosphorylated histone H2AX.