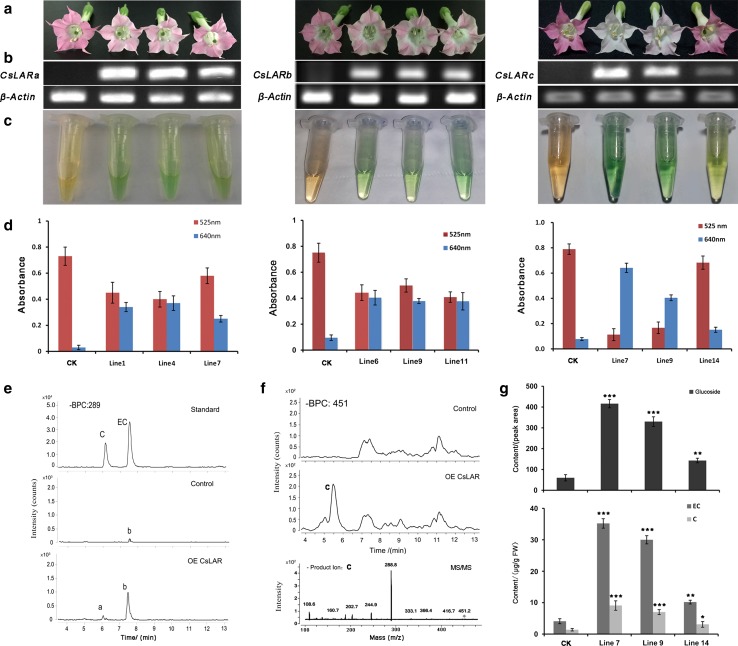
Fig. 6.
Overexpression of CsLARs in wild-type tobacco. a Phenotypes of empty vector control (CK) and CsLARs transgenic tobacco flowers. b Semi-quantitative RT-PCR analysis of the CsLAR and the housekeeping gene NtActin transcription levels in total RNA from the flowers. c DMACA staining of the extracts from transgenic tobacco flowers and CK. d The absorbance of the anthocyanins extracts and DMACA-reactive compounds at 525 and 640 nm, respectively. e Extraction ion chromatogram of products (m/z 289.0) accumulated in the transgenic tobacco flowers and CK. Authentic standard of catechin and epicatechin (top); products accumulated in the transgenic tobacco flowers (bottom) and CK (middle). f Extraction ion chromatogram of products (m/z 451.0) in the transgenic tobacco flowers (bottom) and CK (top). UPLC–MS/MS of peak c (m/z 451.0) is listed in the box. g The contents of glycoside, C and EC extracted from different transgenic lines of overexpressing CsLARc tobacco flowers and CK. The content of glycoside is indicated by the peak area as determined by UPLC–MRM-MS. The contents (mg/g, FW) of C and EC were quantified according to the peak area based on the standard C and EC. All data are the means of three biological replicates, and the error bars represent the standard deviation of three replicates. The asterisks indicate the significance level (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001) based on a Tukey’s honestly significant difference test