Abstract
Nuclear pore complexes (NPCs) are multiprotein channels that bridge the nucleus with the cytoplasm and regulate all nucleo‐cytoplasmic traffic. NPCs are built by the repetition of ~30 different proteins known as nucleoporins (Nups). Accumulating evidence has revealed a diversity in NPC composition that is critical for cell‐specific functionality and fate determination. A new report by Hazawa et al 1 now identifies the central transport channel nucleoporin Nup62 as a novel regulator of cell proliferation and differentiation in squamous cell carcinoma (SCC), via modulation of p63 nucleo‐cytoplasmic transport. These findings provide further evidence on how alterations in NPC composition might be utilized to determine cell fate.
Subject Categories: Cancer, Membrane & Intracellular Transport
In eukaryotic cells, the genome is enclosed by the nuclear envelope, which provides a physical barrier between the cytoplasm and the nuclear interior. All transport in and out of the nucleus occurs through large (~110 MDa), aqueous transport channels that perforate the nuclear envelope at sites where the inner and outer nuclear membranes are fused 2. These channels, called nuclear pore complexes (NPCs), are highly organized structures composed of a membrane‐embedded scaffold which surrounds a central transport channel, a cytoplasmic ring from which filaments project outward into the cytoplasm, and a nuclear ring from which filaments protrude into the nucleoplasm and further connect with a distal ring, forming a structure called the nuclear basket. NPCs are composed of multiple copies of ~30 different proteins, termed nucleoporins (Nups), which assemble to form subcomplexes and localize to the different structural parts of the pore.
Traditionally considered to be static complexes of ubiquitous composition, evidence accumulated over the last few years has shown that NPCs are in fact dynamic structures, with a composition that varies between different tissues, cell types, and developmental stages 3. In addition, mutations of several nucleoporins have been found to be associated with tissue‐specific diseases, suggesting a NPC compositional diversity that is associated with the regulation of specific cell functions 3. Consistent with this idea, NPCs are emerging as important determinants of cell fate. D'Angelo et al 4 first reported that the addition of the tissue‐specific nucleoporin Nup210 to nuclear pores is required for muscle and neuronal differentiation, indicating that this single modification of nuclear pore composition is a key determinant of cell fate. Jacinto et al 5 then reported a role for the nucleoporin Nup153, which localizes to the nuclear basket and also exists outside of the pore in the nucleoplasm, in the regulation of ESC pluripotency. Reduction of Nup153 expression in ESCs was shown to induce early neuronal differentiation, revealing a role for this nucleoporin in maintaining ESC stemness 5. These observations were recently confirmed by Toda et al, 6 who further showed that neuroprogenitor cells require high expression of Nup153 for their maintenance.
New findings from the Wong Lab now expand on these observations by uncovering a role for the central channel NPC component, Nup62, in regulating the proliferation and differentiation state of squamous cell carcinoma (SCC) cells 1. In an initial analysis of nucleoporin gene expression across several healthy tissues, Hazawa et al show that Nup62 is among a subset of Nups that is expressed at higher levels in stratified epithelia of the skin and esophagus. Intriguingly, most of these Nups are also overexpressed in primary SCC samples from both head and neck, as well as cervix cancers, with Nup62 showing the highest levels of expression. siRNA‐mediated reduction of this nucleoporin in several SCC cell lines reduces cell growth and increases the expression of genes associated with differentiation, suggesting a role for Nup62 in maintaining the proliferative state and/or viability of SCC cells. A microarray analysis of genes that are significantly downregulated in Nup62‐depleted cells revealed a strong correlation with genes that are altered by depletion of the p53 homolog p63. These findings are interesting because p63 is critical for the development of stratified squamous epithelium, where it governs self‐renewal, and it is also frequently amplified in SCC 7, 8. The transcriptional regulation of p63 is complex, with distinct promoters leading to transactivational (TAp63) or non‐transactivational (∆Np63) p63 isotypes, each yielding several alternatively spliced isoforms. Of these, ∆Np63α is the predominant isotype within the stratified epithelium 8. Nup62 knockdown in SCC cells reduces nuclear accumulation of ∆Np63α as well as transcription of a subset of its target genes, without affecting p63 mRNA or protein levels, suggesting a role for Nup62 in regulating ∆Np63α nuclear transport. Notably, the authors report a physical association between Nup62 and ∆Np63α and that depletion of Nup62 partially reduces the nuclear levels of ∆Np63α. Through analysis of microarray data from Nup62 knockdown cells, the authors reveal a strong correlation with genes regulated by the differentiation‐inducible kinase ROCK. Inhibition of this serine/threonine kinase has previously been shown to increase ∆Np63α expression in keratinocytes and augment self‐renewal 9. The authors show that ROCK phosphorylates Nup62 on two residues in its phenylalanine–glycine (FG) repeat domain and that this phosphorylation negatively regulates the association between Nup62 and ∆Np63α. Upon phosphorylation, Nup62 dissociates from ∆Np63α, resulting in reduced nuclear import of this transcription factor. Conversely, a Nup62 mutant that cannot be phosphorylated by ROCK increases the expression of ∆Np63α target genes, in addition to increasing SCC proliferation. Altogether, the work by Hazawa et al 1 uncovers a novel mechanism that promotes the proliferation of SCC cells and prevents their differentiation through regulation of ∆Np63α nuclear transport by the NPC component Nup62 (Fig 1). What remains to be determined is whether increased expression of Nup62 is sufficient to drive ∆Np63α nuclear accumulation, and whether the high levels of Nup62, or any of the other nucleoporins upregulated in SCC, contribute to tumor formation and/or maintenance. Also, Nup62 is a component of the nuclear pore central channel and, thus, it likely regulates the transport of many other molecules in SCC that remain to be identified. Finally, in this work cell proliferation was determined exclusively with an assay that measures metabolic activity, which, combined with the observed alterations in cell viability, opens the possibility that Nup62 might regulate SCC metabolism and viability, instead of cell proliferation.
Figure 1. Nup62 prevents SCC differentiation by modulating ∆Np63α nuclear transport.
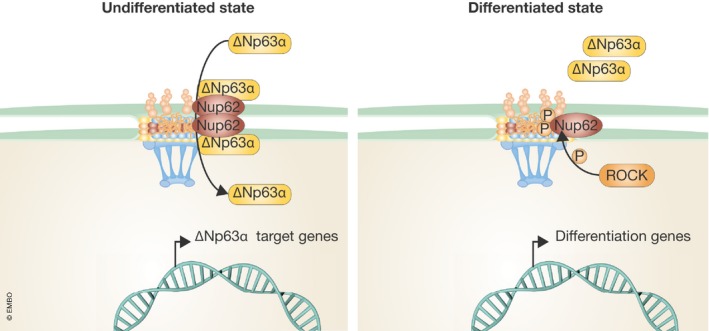
In undifferentiated SCC cells, the NPC component Nup62 interacts with the transcription factor ∆Np63α to mediate its nuclear import. Once inside the nucleus, ∆Np63α drives the expression of genes that maintain the proliferative capacity and stemness of SCCs. Upon differentiation, ROCK phosphorylates Nup62 in two FG domain residues, reducing the association of Nup62 with ∆Np63α. This impairs ∆Np63α nuclear import, which subsequently causes expression of differentiation genes 1.
Previous nucleoporins implicated in cell fate determination were shown to do so via regulation of gene transcription. While Nup210 is essential for assembling a MEF2C‐containing transcription complex at the nuclear periphery of muscle cells 4, 10, Nup153 recruits PRC1 complexes and associates with Sox2 to repress ESC differentiation genes 5, 6. In this study, Hazawa et al report the regulation of cell proliferation and differentiation via modulation of the canonical function of nuclear pore complexes, the regulation of nucleo‐cytoplasmic transport. The work by the Wong group uncovers a novel function for Nup62 and further advances our understanding of how differential expression of NPC components might regulate cell fate.
See also: M Hazawa et al (January 2018)
References
- 1. Hazawa M, Lin D, Kobayashi A et al (2018) EMBO Rep 19: e44523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beck M, Hurt E (2017) Nat Rev Mol Cell Biol 18: 73–89 [DOI] [PubMed] [Google Scholar]
- 3. Raices M, D'Angelo MA (2012) Nat Rev Mol Cell Biol 13: 687–699 [DOI] [PubMed] [Google Scholar]
- 4. D'Angelo MA, Gomez‐Cavazos JS, Mei A et al (2012) Dev Cell 22: 446–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacinto FV, Benner C, Hetzer MW (2015) Genes Dev 29: 1224–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Toda T, Hsu JY, Linker SB et al (2017) Cell Stem Cell 21: 618–634 e617 [DOI] [PubMed] [Google Scholar]
- 7. Yang A, Schweitzer R, Sun D et al (1999) Nature 398: 714–718 [DOI] [PubMed] [Google Scholar]
- 8. Deyoung MP, Ellisen LW (2007) Oncogene 26: 5169–5183 [DOI] [PubMed] [Google Scholar]
- 9. Suprynowicz FA, Upadhyay G, Krawczyk E et al (2012) Proc Natl Acad Sci USA 109: 20035–20040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raices M, Bukata L, Sakuma S et al (2017) Dev Cell 41: 540–554 e547 [DOI] [PMC free article] [PubMed] [Google Scholar]


