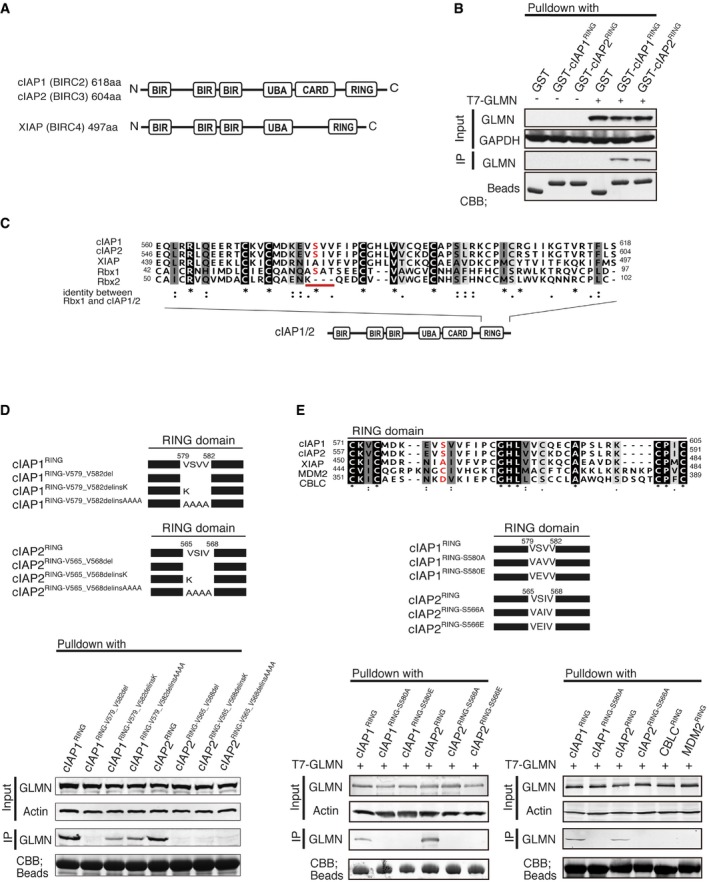
A schematic representation of cIAP1 (BIRC2, 618 aa) and cIAP2 (BIRC3, 604 aa), and XIAP (BIRC4, 497 aa).
GST‐pulldown assays revealed that GLMN binds to the RING domain of cIAPs.
Comparison of the RING‐domain amino acids of Rbx1, Rbx2, cIAP1, cIAP2, and XIAP. Amino acids that are identical (black) or similar amino acids (gray) among the five sequences are shaded. cIAP1 and cIAP2 have a serine residue in the red‐underlined motif that is present in Rbx1 but not Rbx2 or XIAP.
Point‐mutation analysis indicated that the VSV(I)V motif of the cIAP RING domain is important for the interaction with GLMN. VSV(I)V motifs were deleted or mutated as indicated in the upper panel, and the mutants were subjected to pulldown assays.
Upper panel: RING domain sequences were compared with those of two other E3 ligases, MDM2 and CBLC. An amino acid sequence alignment of the RING domains of cIAP1, cIAP2, XIAP, MDM2, and CBLC is shown. MDM2 and CBLC do not have a serine residue in the motif corresponding to the VSV(I)V motif in the cIAPs‐RING domain. Middle panel: schematic representation of point mutations of the serine residue in the VSV(I)V motifs of the cIAPs‐RING domains. Lower panel: MDM2−RING, CBLC−RING, and point‐mutated RING domains were expressed, purified, and subjected to GST‐pulldown assays.
Data information: Data are representative of three similar experiments. See also Fig
.