Salt stress and Pol II-CTD phosphorylation status affect switching between normal snRNA/imRNA production and biogenesis of stress-specific mRNAs derived from 3′-elongated snRNAs/ncRNAs.
Abstract
Phosphorylation of the RNA polymerase II (Pol II) C-terminal domain (CTD) regulates transcription of protein-coding mRNAs and noncoding RNAs. CTD function in transcription of protein-coding RNAs has been studied extensively, but its role in plant noncoding RNA transcription remains obscure. Here, using Arabidopsis thaliana CTD PHOSPHATASE-LIKE4 knockdown lines (CPL4RNAi), we showed that CPL4 functions in genome-wide, conditional production of 3′-extensions of small nuclear RNAs (snRNAs) and biogenesis of novel transcripts from protein-coding genes downstream of the snRNAs (snRNA-downstream protein-coding genes [snR-DPGs]). Production of snR-DPGs required the Pol II snRNA promoter (PIIsnR), and CPL4RNAi plants showed increased read-through of the snRNA 3′-end processing signal, leading to continuation of transcription downstream of the snRNA gene. We also discovered an unstable, intermediate-length RNA from the SMALL SCP1-LIKE PHOSPHATASE14 locus (imRNASSP14), whose expression originated from the 5′ region of a protein-coding gene. Expression of the imRNASSP14 was driven by a PIIsnR and was conditionally 3′-extended to produce an mRNA. In the wild type, salt stress induced the snRNA-to-snR-DPG switch, which was associated with alterations of Pol II-CTD phosphorylation at the target loci. The snR-DPG transcripts occur widely in plants, suggesting that the transcriptional snRNA-to-snR-DPG switch may be a ubiquitous mechanism to regulate plant gene expression in response to environmental stresses.
INTRODUCTION
The eukaryotic transcriptome consists of protein-coding polyadenylated mRNAs and noncoding RNAs (ncRNAs) that have little or no protein-coding potential (Liu et al., 2006). Expression of mRNAs and ncRNAs is regulated by various developmental and environmental stimuli, suggesting unique functions for ncRNAs in growth, development, and stress responses (Brown et al., 1992; Valgardsdottir et al., 2005, 2008; Heo and Sung, 2011). ncRNAs are classified into canonical and noncanonical species; the former includes rRNAs, tRNAs, and small nuclear/nucleolar RNAs (snRNAs and snoRNAs), and the latter includes microRNAs, small interfering RNAs, long ncRNAs, and intermediate ncRNAs (Wang et al., 2014). Early studies established that canonical ncRNAs have roles in protein synthesis and RNA maturation (Lerner et al., 1980; Ohshima et al., 1981; Sontheimer and Steitz, 1993), whereas studies on gene silencing pathways revealed regulatory roles of many noncanonical ncRNAs in gene expression (Ruiz et al., 1998; Reinhart et al., 2002; Xie et al., 2005; Wierzbicki et al., 2009). In Arabidopsis thaliana, some ncRNAs and ncRNA-like sequence have more than one function. For example, tRNA-like structures in mRNAs can function as signals for systemic long-distance transport of the mRNA (Zhang et al., 2016). In addition, the miRNA precursor pri-miR165a functions as an mRNA to produce a regulatory peptide (Lauressergues et al., 2015). Whether or not other classes of ncRNAs can have multiple functions has been unclear. U snRNAs, a major class of canonical ncRNAs, have only been associated with snRNP functions in RNA maturation processes (Lerner et al., 1980; Ohshima et al., 1981; Sontheimer and Steitz, 1993); however, mRNAs with embedded U snRNA structure have been identified in Arabidopsis (Hare et al., 2003).
There are two classes of U snRNAs. Sm-class snRNAs including U1, U2, U4, U4atac, U5, U11, and U12 snRNAs are transcribed by RNA polymerase II (Pol II). These snRNAs have a 5′-trimethylguanosine cap structure and binding sites for Sm proteins (Lerner and Steitz, 1979). LSm-class snRNAs such as U6 and U6atac are transcribed by RNA polymerase III (Pol III). Transcriptional activation of snRNA promoters resembles that of protein-coding gene promoters and requires a conserved snRNA-specific transcriptional activator, SNAPc complex, which binds to proximal sequence element or upstream sequence element (USE) in the promoter (Sadowski et al., 1993; Ohtani and Sugiyama, 2005), and general transcription factors, such as TATA binding proteins, TFIIB, TFIIE, and TFIIF that assemble on TATA-box sequence (Kuhlman et al., 1999). The transcription termination and 3′-end formation mechanisms of snRNA have been studied more in detail in animals and resemble those of mRNA, although they require the presence of a snRNA-specific promoter (Hernandez, 1985; Hernandez and Lucito, 1988). Like pre-mRNAs, pre-snRNAs are transcribed beyond the 3′ end of the mature snRNAs and processed by the Integrator complex that is recruited to Pol II with specific phosphorylation patterns at the C-terminal domain (CTD) of the largest subunit (Egloff et al., 2010). In plants, transcriptions of both classes of snRNAs are regulated by USE (RTCCCACATCG) and TATA box in the promoter, but the spacing between two elements distinguishes them. The Pol II-dependent Sm-class snRNA promoters show [USE-N32-35-TATA box] configuration, whereas Pol III-dependent LSm-like-class promoter shows [USE-N23-24-TATA box] (Waibel and Filipowicz, 1990).
Pol II CTD consists of multiple repeats of a conserved heptapeptide Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (Allison et al., 1988; Nawrath et al., 1990). All residues in heptads except prolines are targets of Pol II-CTD kinases and phosphatases that regulate the activities and functions of Pol II complex during the transcription cycles. Studies in animal systems have shown that the roles of Pol II-CTD phosphoregulation in snRNA transcription largely mirror those in mRNA transcription (Egloff, 2012; Egloff et al., 2012; Wani et al., 2014). During transcription activation and promoter escape, phosphorylation at Ser5 is required to recruit capping enzymes (Ho and Shuman, 1999; Wen and Shatkin, 1999). The Ser2-PO4 mark is crucial for recruiting the Integrator complex during termination and 3′-end processing (Egloff et al., 2010; Gu et al., 2013; Davidson et al., 2014), but not during transcription elongation (Medlin et al., 2003), highlighting differences between transcription of snRNAs and protein-coding genes, at least in animals. Also, Ser7-PO4 phosphorylation is needed for efficient recruitment of the Integrator complex (Egloff et al., 2007, 2012; Egloff, 2012). By contrast, phosphorylation of Ser5 appears inhibitory for the recruitment of the Integrator complex. Information for snRNA transcription in plants is limited, but studies showed that it uses components homologous to animal counterparts such as 11-bp USE (RTCCCACATCG) (Waibel and Filipowicz, 1990), the SNAPc complex (Ohtani and Sugiyama, 2005), and the Integrator complex (Liu et al., 2016). Notable differences between plant and animal snRNA transcription are that plant snRNA 3′-end processing does not require a snRNA promoter (Connelly and Filipowicz, 1993). Involvement of Pol II-CTD phosphorylation in regulation of plant snRNA biogenesis is not clear, but CTD kinase mutants with decreased CTD phosphorylation levels did not show decreased snRNA levels in Arabidopsis (Hajheidari et al., 2012).
Phosphorylation status of the Pol II CTD responds to various developmental and environmental signals. For example, human, simian, rodent, and avian cells treated with severe heat shock exhibit higher Pol II-CTD phosphorylation, whereas amphibian and insect Pol II apparently reduce the CTD phosphorylation level after heat shock (Lavoie et al., 1999). Similarly, heat stress has been shown to elevate Ser2 phosphorylation level in budding yeast (Saccharomyces cerevisiae; Patturajan et al., 1998). On the other hand, mild heat stress causes dephosphorylation of Pol II CTD due to CTD-kinase inhibition in human cells (Dubois et al., 1994; Venetianer et al., 1995). In Drosophila melanogaster, Pol II-CTD dephosphorylation by the TFIIF-associating CTD-phosphatase 1 (FCP1) is required for efficient transcription of heat shock genes (Fuda et al., 2012). DmFCP1 depletion by RNA interference (RNAi) did not cause significant changes in Pol II-CTD phosphorylation level on heat shock genes. Instead, it caused a dramatic increase of non-chromatin-bound hyperphosphorylated Pol II. In the fission yeast Schizosaccharomyces pombe, nitrogen starvation promotes MAP kinase Sty1 to phosphorylate Pol II-CTD kinase CTDK-I and induces CTD Ser2 phosphorylation (Sukegawa et al., 2011). Osmotic stress and oxidative stress have been shown to give rise to a “Pol IIm” form, which is phosphorylated extensively on Ser5 due to ERK/MAPK activities (Bonnet et al., 1999). In Arabidopsis, CTD phosphoregulation is an integral part of various biotic and abiotic stress signaling including microbial-associated molecular patterns/innate immunity signaling (Li et al., 2014), osmotic stress response (Koiwa et al., 2002; Xiong et al., 2002), and hormone signaling (Ueda et al., 2008; Matsuda et al., 2009). At an ecological scale, the allelopathic phytochemical juglone (5-hydroxy-1,4-naphthalenedione) inhibits a peptidyl-prolyl isomerase Pin1 and causes dephosphorylation of Pol II CTD in animal cell models, suggesting that CTD is a target of allelopathic interactions (Hennig et al., 1998; Chao et al., 2001).
CTD-PHOSPHATASE-LIKE4 (CPL4) is a CTD phosphatase in Arabidopsis and is orthologous to human and fungal FCP1 (Fukudome et al., 2014). Knocking down CPL4 expression by RNAi (CPL4RNAi) induces global CTD hyperphosphorylation and upregulation of more than 200 genes, indicating that CPL4 functions as a major CTD phosphatase in Arabidopsis. In this study, we show that CPL4RNAi also affect snRNA biogenesis, promoting 3′-extensions of snRNA transcripts. Surprisingly, this resulted in production of chimeric snRNA-fused mRNAs with protein-coding potential. Expression of these fusion transcripts, which are termed snR-DPG (downstream protein-coding gene) transcripts, is dependent on snRNA transcription machinery. snR-DPG transcripts are also produced in the wild-type background when CTD phosphorylation status is altered by salt stress and detected in a specific tissue. These results revealed a key mechanism of plant stress-responsive gene expression that uses Pol II-CTD phosphorylation and snRNA 3′-end processing as regulatory nodes.
RESULTS
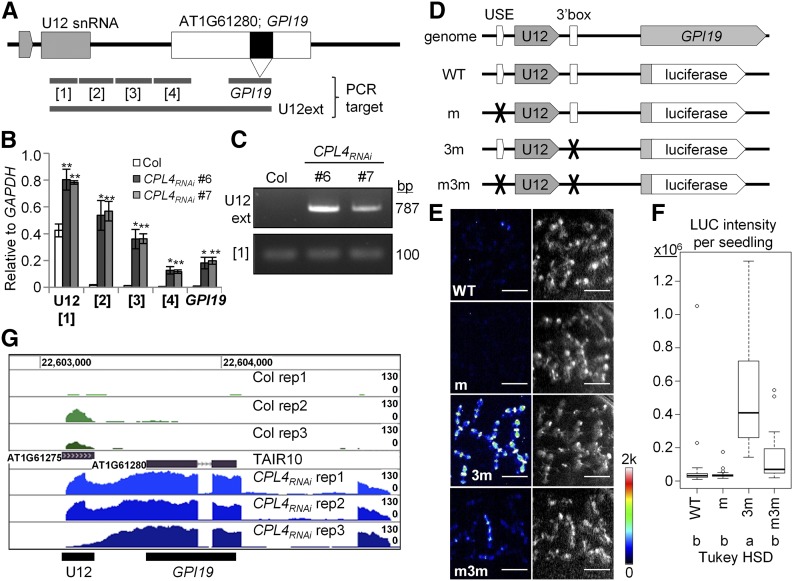
Upregulation of AT1G61280 (GPI19) in CPL4RNAi Lines Is Coupled with a 3′-Extension from an Upstream U12 snRNA Gene
Our previous microarray study using CPL4RNAi Arabidopsis ecotype Columbia gl1 (Col) detected three major coregulated clusters of genes affected by silencing CPL4 expression, involved in xenobiotic stress responses, abscisic acid responses, and flavonoid biosynthesis pathway clusters (Fukudome et al., 2014). Interestingly, the most upregulated gene in CPL4RNAi (AT1G61280: putative GPI19/PIG-P subunit) did not show any coregulation with the major gene clusters (Fukudome et al., 2014). The ESTs (GenBank accession numbers DR367811.1 and DR384064.1) for GPI19 revealed a unique transcript structure containing a whole U12 snRNA in the 5′ untranslated region of GPI19. Apparently, transcription of GPI19 can originate from a transcription start site of U12 snRNA gene (AT1G61275) located 290 bp upstream of GPI19. This suggested that upregulation of GPI19 in CPL4RNAi is due to alteration of U12 snRNA transcription regulation.
To test if CPL4RNAi indeed produced a U12-GPI19 fusion transcript, we conducted RT-qPCR analysis of regions between U12 snRNA and GPI19 (Figures 1A to 1C). In the wild type, the only transcript detected in this region is mature U12 snRNA (Figure 1B). By contrast, in CPL4RNAi, transcripts were detected throughout the intergenic region and GPI19 coding region. Also, the continuous U12-GPI19 transcript was detected by RT-PCR (Figure 1C). These results suggested that CPL4 regulates expression of GPI19 via a nonconventional regulatory mechanism, extending 3′ end of upstream snRNA transcripts.
Figure 1.
Upregulation of GPI19 in CPL4RNAi Is Coupled with 3′-Read-Through of an Upstream U12 snRNA Gene.
(A) Structure of genomic region containing U12 snRNA and GPI19 (AT1G61280) genes (gray and white boxes, respectively). A black box within GPI19 indicates an intron. Gray bars show RT-qPCR target regions used in (B) and (C).
(B) Expression level of each fragment described in (A) relative to GAPDH, measured by RT-qPCR. Error bars indicate se of the mean of biological triplicates. *P < 0.05 and **P < 0.01 by one-tailed Student’s t test between Col and each CPL4RNAi line.
(C) RT-PCR analysis detecting the 3′-extended U12 snRNA.
(D) Design of the U12-LUC reporter system. Luciferase is translationally fused to the GPI19 N-terminal fragment.
(E) Luminescence (left) and bright- field (right) images of 1-week-old U12-LUC reporter lines. A representative line with median LUC intensity was chosen for each construct. Exposure time was 20 min.
(F) Box plot showing mean luciferase intensity per seedling of independent U12-LUC lines. Numbers of independent lines examined are 21, 24, 17, and 24 for the wild type, m, 3m, and m3m, respectively. For each line, 18 to 32 seedlings were counted. Different letters in Tukey HSD test indicate significant differences between genotypes (one-way ANOVA followed by Tukey HSD test, P < 0.05).
(G) RNA-seq read coverage over the U12-GPI19 locus in Col and CPL4RNAi lines. Coverage values were normalized by library (total amount of mapped read) and associated bigWig track formats for visualization in Trackster viewer were generated by BEDTools and Wig/BedGraph-to-bigWig converter, respectively.
A 3′-Box Mutation in U12 Enhances Downstream Protein-Coding Gene Expression
To test if the U12-GPI19 fusion transcript is translatable like a protein-coding mRNA, we prepared a reporter gene using a 1467-bp genomic fragment, spanning from 981 bp upstream of U12 to 22 bp downstream of the ATG codon of GPI19, fused to the firefly luciferase (LUC) coding region (Figure 1D). Furthermore, the roles of U12 USE and 3′-box were tested by introducing mutations in the reporter gene. In the Arabidopsis ecotype Col-0 host plants, the LUC activity from the wild-type reporter was detectable but weak. By contrast, the 3m reporter with compromised 3′-box showed the highest LUC activities, which were abolished by additional mutations in USE (m3m; Figures 1D to 1F; Supplemental Table 1). These results show that the U12-LUC expression is dependent on the snRNA promoter and that read-through of snRNA 3′-box can enhance expression of the downstream protein-coding gene.
The impact of CPL4RNAi on U12-LUC reporter genes was tested by crossing the reporter lines to CPL4RNAi. Interestingly, CPL4RNAi showed high induction of U12-LUC reporter genes with functional USE (Table 1), showing 140- and 52-fold induction for the wild-type and the 3m variant reporter genes, respectively. On the other hand, CPL4RNAi did not substantially enhance the expression levels of the U12-LUC with the USE mutation (m and m3m; fold induction 6 and 2, respectively). Notably, the expression level of wild-type U12-LUC in CPL4RNAi was higher than 3m variants in Col-0 background, indicating that CPL4RNAi promotes not only read-through of snRNA 3′-box, but also the continuation of read-through transcription at the downstream of 3′-box. Overall, the results suggested that part of U12 snRNA transcription was switched to produce U12-GPI19 mRNA in CPL4RNAi.
Table 1. Effects of CPL4RNAi on U12-LUC Reporter Expression.
| U12-LUC Genotype | n | LUC Activity per Seedling |
P Value (t Test) | |
|---|---|---|---|---|
| Ave. | se | |||
| U12_WT | 18 | 7.76 × 104 | ± 9.90 × 103 | – |
| U12_WT × CPL4RNAi F1 | 13 | 1.09 × 107 | ± 9.21 × 105 | 6.02 × 10−8 |
| U12_3m | 14 | 2.59 × 106 | ± 1.61 × 105 | – |
| U12_3m × CPL4RNAi F1 | 10 | 1.36 × 108 | ± 3.04 × 106 | 1.73 × 10−3 |
| U12_m | 14 | 2.13 × 104 | ± 7.56 × 102 | – |
| U12_m × CPL4RNAi F1 | 8 | 1.31 × 105 | ± 1.13 × 104 | 2.50 × 10−5 |
| U12_m3m | 17 | 8.63 × 104 | ± 1.02 × 104 | – |
| U12_m3m × CPL4RNAi F1 | 17 | 1.91 × 105 | ± 1.54 × 104 | 2.95 × 10−6 |
LUC activities in individual F1 seedlings from the cross between U12-LUC transgenic lines and the CPL4RNAi line were quantified. See Figure 1E for the reporter gene designs. P values are from two-tailed Student’s t test between the parental and F1 lines.
Pol II-Dependent snRNA Loci Show 3′-Extension in CPL4RNAi Lines
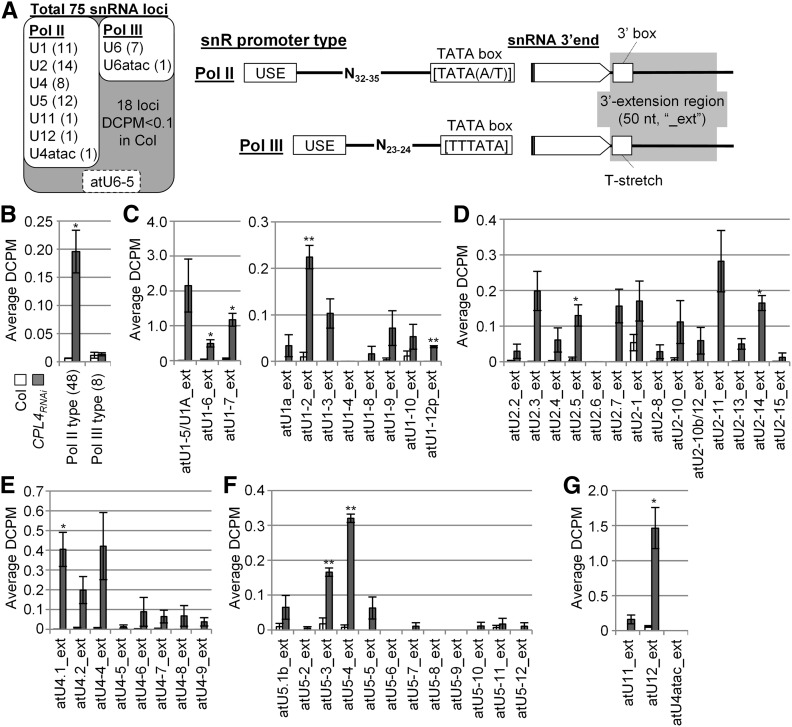
To test if CPL4RNAi induces 3′-extensions of other snRNAs, we conducted RNA-seq analysis of the CPL4RNAi transcriptome. Sequencing libraries containing cDNA from both mRNAs and ncRNAs were prepared using the rRNA depletion kit. As shown in Figure 1G, a high level of RNA-seq reads was continuously mapped to the region spanning U12 and GPI19, consistent with the RT-PCR analyses. The reads coverage spans the entire GPI19 coding region and 3′ untranslated region, suggesting that this transcript is translatable. Of all 75 snRNA loci in the TAIR10 reference genome (Wang and Brendel, 2004), we detected expression of 56 loci in the wild type with depth of coverage per base per million reads mapped (DCPM) higher than 0.1 (Supplemental Data Set 1). Based on the spacing between the USE and the TATA-box in their promoters, 48 and 8 snRNA loci are Pol II and Pol III dependent, respectively (Figure 2A). atU6-5 has an unusual promoter structure and was excluded from the analysis.
Figure 2.
Detection of 3′-Extended snRNA Transcripts in the CPL4RNAi Line.
(A) The expression levels of 75 snRNA loci detected by RNA-seq analyses. The DCPM was determined for each snRNA locus. Among those, 56 loci with DCPM higher than 0.1 in Col (white box) were selected for further analysis. RNA polymerase specificity was determined for each locus based on the spacing (Pol II, 32–34 nucleotides; Pol III, 23–24 nucleotides) between USE and TATA box. The 3′-box signal and T stretch indicate snRNA 3′-processing signal for Pol II and Pol III, respectively. A U6 snRNA, atU6-5, has a noncanonical spacing between USE and TATA box and lacks T-stretch signal in its 3′-end, thus was removed from further analysis.
(B) to (G) The depth of coverage of the 3′-extended regions of 56 snRNAs selected in CPL4RNAi. All error bars indicate se of the mean of biological triplicates. *P < 0.05 and **P < 0.01 by two-tailed Student’s t test between Col and CPL4RNAi.
(B) Average DCPM of all snRNA-extensions examined.
(C) to (G) U1 snRNAs (C), U2 (D), U4 (E), U5 (F), and U11, U12, and U4atac (G) minor snRNAs.
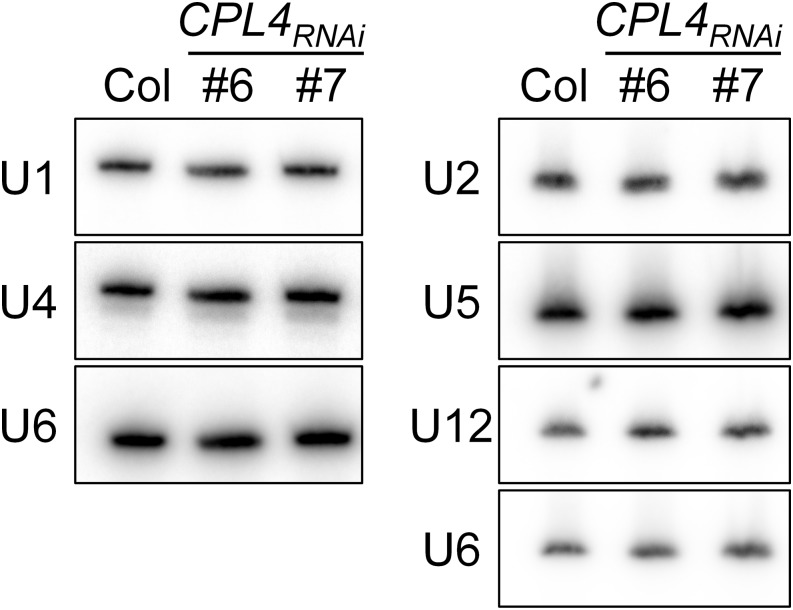
To measure transcript extension, we determined levels of transcript mapped downstream (50 nucleotides) of each of the 56 snRNA loci (Figure 2A, right, highlighted in gray). The analysis revealed that CPL4RNAi shows significantly higher coverage in the 3′-extension regions of all classes of Pol II-dependent snRNAs, but not in those of Pol III-dependent U6 snRNA loci (Figures 2B to 2F; Supplemental Data Set 1). RT-PCR analysis of selected snRNA loci confirmed RNA-seq data (Supplemental Figure 1). On the other hand, the levels of mature snRNAs in the wild type and CPL4RNAi were similar to each other for Pol II- and Pol III-dependent snRNAs (Figure 3). These results showed that CPL4RNAi globally promoted accumulation of 3′-extended, read-through transcripts for snRNAs. By contrast, CPL4RNAi did not show clear promotion of 3′-extensions of protein-coding mRNAs. In a similar analysis using 9003 protein-coding loci with detectable transcripts, we detected 45 and 33 loci (Student’s t test, P < 0.01) with increased and decreased mRNA 3′-extension, respectively, in CPL4RNAi. Considering a small number of detected genes and no clear increase or decrease in the levels of the mRNA 3′-extensions, we concluded that the transcript 3′-extension in CPL4RNAi is specific to Pol II-dependent snRNA loci.
Figure 3.
RNA Gel Blot Analysis of Mature snRNAs from Col and CPL4RNAi Lines.
Total RNA was extracted from 7-d-old seedlings and separated on 6% polyacrylamide gels. Amount of RNA loaded to each well were 1.5 and 10 µg for the left and right panels, respectively.
Chimeric snR-DPG Transcripts Accumulate in CPL4RNAi Lines
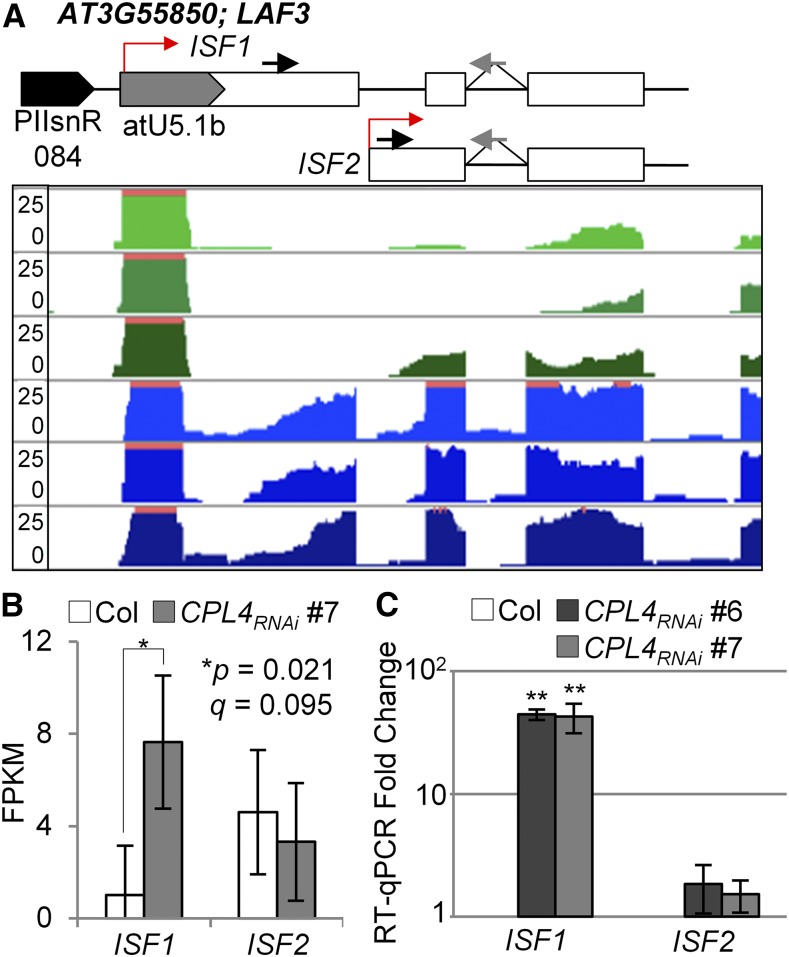
Among the 48 Pol II-dependent snRNAs examined, 18 have a DPG within 1 kb of the annotated snRNA (Table 2). The majority of the identified DPGs are upregulated in the CPL4RNAi line (Table 2). The coding sequences of the DPGs are properly spliced, as exemplified by the U2.5-AT5G09590 chimeric transcript (Supplemental Figure 1B). To test if upregulation of DPGs in CPL4RNAi was indeed due to the snRNA extension or to activation of an overlapping mRNA promoter, we conducted a detailed characterization of the LONG-AFTER FAR-RED3 (LAF3) locus (AT3G55850). Notably, the LAF3 locus produces two mRNA isoforms with or without embedded snRNA. Isoform1 (LAFISF1; GenBank accession number AY295343.1, BX823543.1) transcription starts with the at U5.1b snRNA, whereas isoform2 (LAFISF2) starts at 223 bp downstream of the U5 snRNA sequence (Figure 4A), likely representing functions of distinct snRNA promoter (ISF1) and mRNA promoter (ISF2). Isoform-specific RT-qPCR analysis showed that in the CPL4RNAi background, the chimeric LAF3ISF1 specifically overaccumulated without an upregulation of LAF3ISF2 (Figures 4B and 4C), indicating that at least in the U2.5-LAF3 locus, the overexpression of DPG in CPL4RNAi is specifically due to the snRNA extension.
Table 2. Eighteen Pol II-Dependent snRNA Loci with Proximal DPG on the Same Strand.
| snRNA |
Space (bp) | DPG |
||||
|---|---|---|---|---|---|---|
| snRNA | snR-3′ext Level | AGI | Gene Description | RNA-Seq FC (Log2) | q-Value | |
| atU12 | High | 298 | AT1G61280 | GPI19/PIG-P subunit | 6.30 | 0.11 |
| atU2-11 | Moderate | 120 | AT1G09800 | Pseudouridine synthase family protein | 2.58 | 0.01 |
| atU1-2 | Moderate | −170 | AT4G23420 | NAD(P)-binding Rossmann-fold superfamily protein | −2.42 | 0.00 |
| atU2-1 | Moderate | 272 | AT1G16825 | Reticulon family protein | 0.72 | 0.10 |
| atU5-3 | Moderate | 689 | AT1G70190 | Ribosomal protein L7/L12 | 0.28 | 0.32 |
| atU2.7 | Moderate | 463 | AT5G61450 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | 0.54 | 0.02 |
| atU2.5 | Moderate | 340 | AT5G09590 | HSC70-2 | 1.05 | 0.00 |
| atU1-3 | Moderate | 396 | AT5G51680 | Hydroxyproline-rich glycoprotein family protein | n/a | 1.00 |
| atU4-6 | Low | 129 | AT1G11870 | Seryl-tRNA synthetase (SRS) | −0.30 | 0.14 |
| atU5.1b | Low | −116 | AT3G55850 | LAF3ISF2 | 0.96 | 0.00 |
| atU2.4 | Low | 61 | AT3G56820 | – | 0.89 | 0.07 |
| atU2-13 | Very low | 60 | AT2G20410 | RNA-binding ASCH domain protein | 0.65 | 0.03 |
| atU4-9 | Very low | 802 | AT1G79970 | – | −0.13 | 0.56 |
| atU2-8 | Very low | 455 | AT5G67560 | ATARLA1D; ADP-ribosylation factor-like A1D | 0.11 | 0.61 |
| atU4-5 | Very low | 337 | AT5G25770 | α/β-Hydrolase superfamily protein | 0.45 | 0.29 |
| atU5-7 | Very low | 476 | AT4G02530 | – | 0.35 | 0.04 |
| atU5-6 | nd | 1045 | AT1G04470 | Unknown function (DUF810) | n/a | 1.00 |
| atU5-9 | nd | 854 | AT1G79540 | – | 0.20 | 0.45 |
The snR-3′ext level is defined as snR-ext region DCPM > 1.0, high; 1.0 > DCPM > 0.1, moderate; 0.1 > DCPM > 0.05, low; DCPM < 0.05, very low; no 3′-extension, nd. snR-DPG spacing column shows distance in base pairs between the last base of snRNA and the first base of DPG. Negative values indicate the snRNA is embedded in the DPG. Fold-change (log2) of DPG and associated q-values were obtained by Cuffdiff.
Figure 4.
Expression of U5 snRNA-Fused LAF3ISF1 in CPL4RNAi.
(A) RNA-seq mapping coverage tracks of the LAF3 genomic region for Col (green) and CPL4RNAi (blue). Location of snRNA promoter (PIIsnR) and transcript isoforms and primers for isoform-specific RT-qPCR analysis are shown on top. Open boxes indicate exons. Red arrows indicate transcription start sites. Black and gray arrows show forward and reverse primer positions for qPCR (Supplemental Data Set 4).
(B) FPKM (fragments per kilobase million) of ISF1 (tss_id; TSS902) and ISF2 (tss_id; TSS35900) in the wild type and CPL4RNAi, and associated P and q-values computed by Cuffdiff are shown. Error bars show confidence intervals from biological triplicates.
(C) The increase of ISF1 expression in CPL4RNAi lines measured by RT-qPCR. Forward primers designed to detect specific isoform and a reverse primer spanning 2nd intron-exon junction were used. Mean fold-change values relative to Col are plotted in log10 scale, with error bars showing se of the mean of biological triplicates. **P < 0.01 by two-tailed Student’s t test between Col and each CPL4RNAi line.
A Transposon-Embedded PIIsnR Drives the AT1G20320 (SSP14) Locus to Produce a 3′-Extension-Capable Short Unstable Intermediate-Length RNA
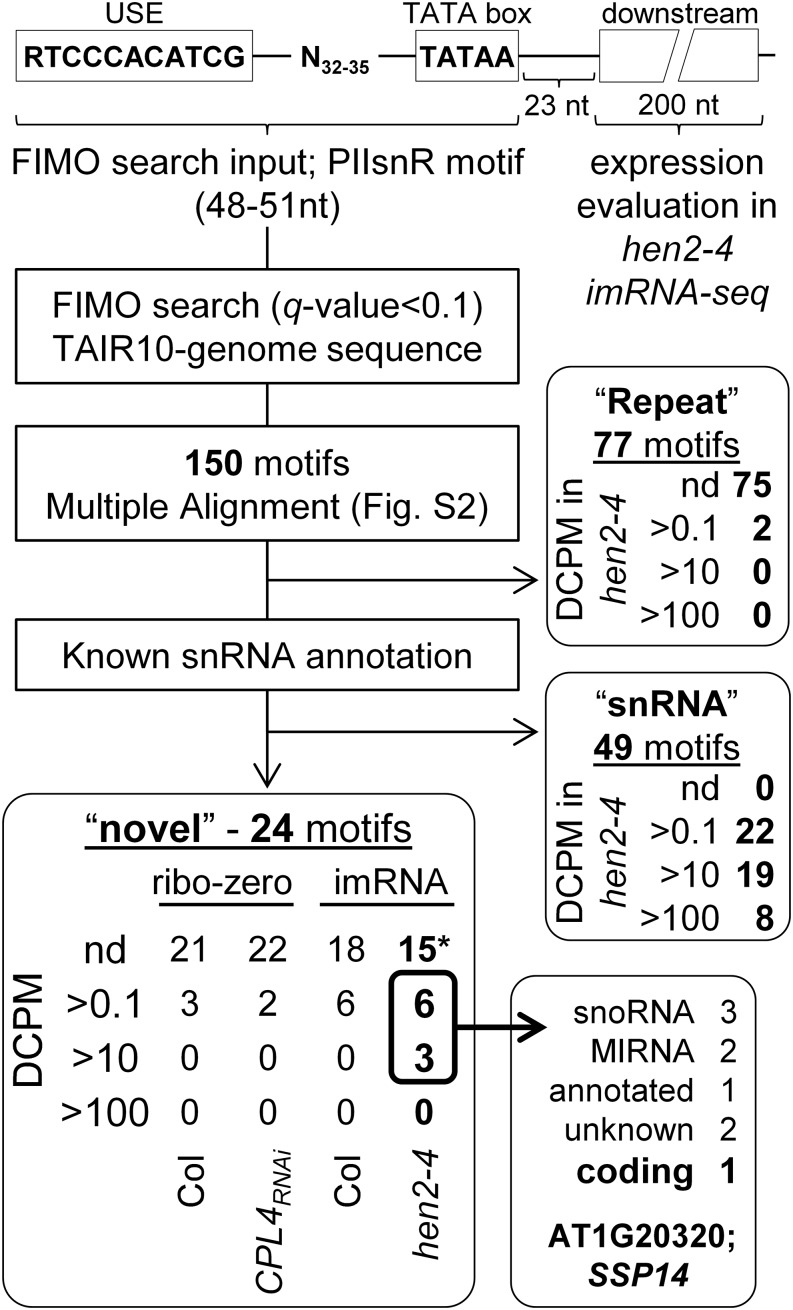
The above results suggested that CPL4 regulates expression of a suite of protein-coding genes that are driven by Pol II-dependent snRNA promoters (PIIsnR). Next, we tested if there are protein-coding genes that lack snRNA sequences but are regulated by PIIsnR. A genome-wide search identified 150 PIIsnR motifs (RTCCCACATCG-N32-35-TATAA) in Arabidopsis (Figure 5; Supplemental Data Set 2). Of these, 77 highly conserved motifs were part of repeat sequences related to transposable elements (TEs) and therefore were classified as “repeat” sequences. Most of the identified TEs were helitron-type nonautonomous DNA transposons or their fragments (Supplemental Figure 2; Table 3). Indeed, PIIsnR sequences could be found in the consensus sequence of ATREP5 and HELITRONY1D in Arabidopsis (Bao et al., 2015) and in those of LTR and Gypsy from other green plants (Table 3). Forty-nine “snRNA” motifs were located upstream of known snRNA loci. The remaining 24 motifs were classified as “novel,” which included one protein-coding gene AT1G20320 (SMALL SCP1-LIKE PHOSPHATASE14 [SSP14]). Expression levels of these loci were inspected in RNA-seq data (ribo-zero protocol) of Col plants. Most of the “repeat” or “novel” loci did not produce significant levels of transcripts except three snoRNAs in the “novel” class (Figure 5). One possible reason for this observation was that the standard RNA-seq strategy was not effective for the detection of relatively short and unstable transcripts. To improve the detection efficiency, the RNA-seq strategy was modified. First, to stabilize short-lived RNA, we used a HUA ENHANCER2 mutant (hen2-4), which is defective in the nuclear exosome pathway for RNA degradation (Lange et al., 2014). Second, the RNA-seq protocol was modified to sequence intermediate-length RNA (imRNA). The imRNA sequencing protocol improved detection of PIIsnR-driven transcripts; in particular, the imRNA profile for hen2-4 revealed production of transcripts from 9 out of 24 “novel” loci (Figure 5, Table 4). These loci include three monocistronic snoRNAs (Marker et al., 2002), two MIRNAs, a long ncRNA (AT4NC021500), and a protein-coding gene, SSP14.
Figure 5.
Workflow of Bioinformatic Analysis to Identify the Genome-Wide Distribution of Pol II-Dependent snRNA Promoter Motifs in Arabidopsis.
A motif containing two core promoter elements for snRNA transcription (USE and TATA box) spaced by 32- to 35-nucleotide sequence was used as a query in FIMO search against Arabidopsis TAIR10 genome. The downstream 200-nucleotide region starting at 24 nucleotides downstream of the TATA box was used to evaluate the expression levels (DCPM) of each locus in RNA-seq data sets, and the numbers of PIIsnR with an indicated level of expression are shown. *PIIsnR090, 039, and 070 in “novel” category showed average DCPM higher than 0.1, but reads were mapped on the opposite strand, therefore were considered as not detected (nd).
Table 3. Consensus Sequences of Green Plant TEs Bearing a PIIsnR Motif.
| Sequence Name | Species | TE | Str | Start | q-Valuea | Matched Sequence (RTCCCACATCG-N32-35-TATAA) |
|---|---|---|---|---|---|---|
| Gypsy-140_SBi-LTR | Sb | LTR | + | 468 | 1.4E-03 | GTCCCACATCGcctgtccagaagaggtgggaggcttttctaggcTATAA |
| Gypsy-29_Mad-LTR | Md | LTR | + | 177 | 2.7E-03 | GTCCCACATCGaccgcggacaaaagctggagactctcctcaacTATAA |
| HARB-1N1_Mad | Md | Harbinger | + | 467 | 3.1E-03 | GTCCCACATCGgctgtgggagaggtttgagcaatcaaacatgctTATAA |
| Gypsy-3_Pru-LTR | Pp | LTR | + | 154 | 6.2E-03 | GTCCCACATCGGaaatatgagcacagtgcacacctcccaaggcctaTATAA |
| Gypsy-5_Pru-LTR | Pp | LTR | + | 179 | 3.3E-03 | GTCCCACATCGgaactttgtgcaaacctcctactttcacctcccTATAA |
| Gypsy-9_AIp-LTR | Ai | LTR | + | 88 | 1.4E-03 | GTCCCACATCGcctaatactcgaaggctccccctcccctactagTATAA |
| Copia-38_JC-LTR | Jc | LTR | + | 19 | 3.7E-03 | ATCCCACATCGaaagaaagggaagggaatagggagttgtttggcTATAA |
| ATREP5 | At | Helitron | + | 1,045 | 1.0E-02 | GTCCCACATCGcttaaaaaaattggacaatggtcaagagccatacTATAA |
| ATREP5 | At | Helitron | + | 481 | 1.0E-02 | GTCCCACATCGcttagaaaaattggacaatggttcagacccatatTATAA |
| HELITRONY1D | At | Helitron | + | 430 | 1.0E-02 | GTCCCACATCGcttaaaaaaattggacattggttcagagccatacTATAA |
| Helitron-6_ALy | Al | Helitron | − | 10,006 | 1.8E-02 | ATCCCACATCGggacggttgactaaaataaatcacttacgtttagaTATAA |
List of consensus sequences of green plant TEs with PIIsnR motifs. Uppercase letters in matched sequence indicate the USE and TATA-box sequences. Str, strand; Sb, Sorghum bicolor; Md, Malus domestica; Pp, Prunus persica; Ai, Arachis ipaensis; Jc, Jatropha curcas; At, Arabidopsis thaliana; Al, Arabidopsis lyrata.
q-value for FIMO search.
Table 4. PIIsnR Motifs in the “Novel” Category with Reads Detected in Their Downstream Region.
| PIIsnR_ID | Str | Chr. | Start | End | Gene/ncRNA Associated with the Motif | Reads Detected in hen2 imRNA-Seq? | Reads Detected in CPL4RNAi RNA-Seq? | 3′-ext Reads Detected in CPL4RNAi RNA-Seq? |
|---|---|---|---|---|---|---|---|---|
| PIIsnR011 | − | 1 | 7,034,923 | 7,034,972 | SSP14; AT1G20320 | Yes | Yes | Yes |
| PIIsnR034 | + | 1 | 23701,098 | 23,701,147 | SNOR146; AT1G08643 | Yes | Yes | Yes |
| PIIsnR134 | + | 5 | 16,598,702 | 16,598,751 | SNOR108; AT5G41471 | Yes | Yes | Weak |
| PIIsnR014 | − | 1 | 10,151,824 | 10,151,872 | SNOR105; AT1G29071 | Yes | Yes | No |
| PIIsnR020 | + | 1 | 13,067,096 | 13,067,145 | MIR773a; AT1G35501 | Yes | No | No |
| PIIsnR095 | + | 4 | 4,824,544 | 4,824,592 | AT4NC021500 | Yes | No | No |
| PIIsnR019 | − | 1 | 13,050,913 | 13,050,960 | MIR773b; AT1G06903 | Yes | No | No |
| PIIsnR002 | + | 1 | 1,131,563 | 1,131,612 | (No similarity)* | Yes | No | No |
| PIIsnR066 | − | 2 | 18,878,387 | 18,878,437 | (No similarity)* | Yes | No | No |
Among 24 PIIsnR motifs categorized as “novel” (not associated with repeat or snRNA), nine show reads mapped to immediately downstream of the motif in the hen2 mutant RNA-seq. See Figure 5 for the search criteria. The PsnR motif’s coordinates, annotations associated with these motifs, as well as hen2 or CPL4RNAi RNA-seq observations, are summarized. Asterisks indicate failure to find a similarity to any sequences by BLAST search. Str, strand.
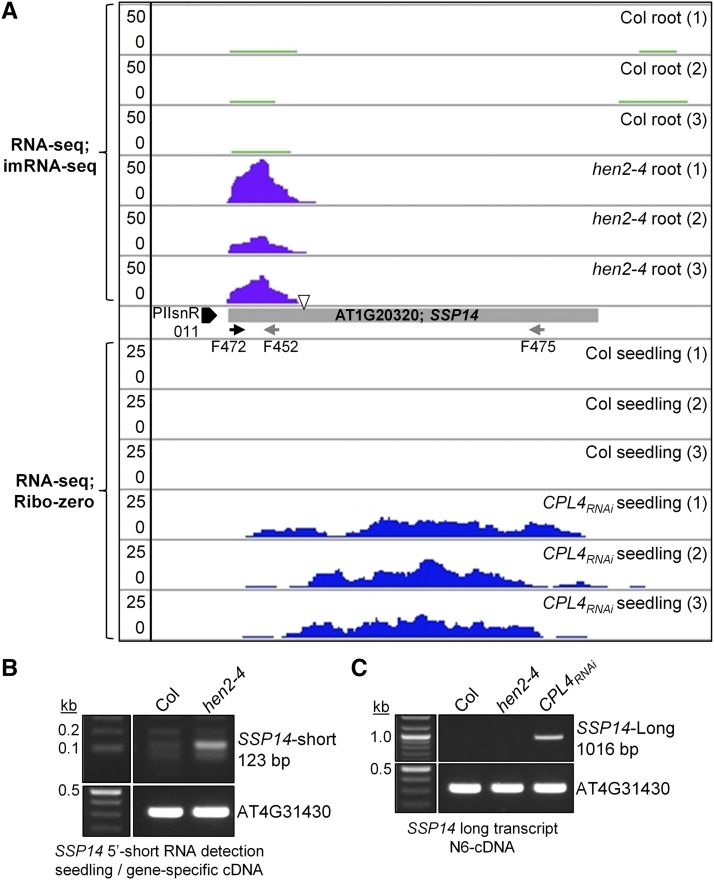
Interestingly, reads mapped to the SSP14 locus in hen2-4 were limited to the 5′-end region (Figure 6). SSP14 is an intronless gene encoding an SCP1-like small phosphatase family protein, SSP14 (Koiwa, 2006). The PIIsnR of SSP14 overlaps with the AtREP5 helitron fragments and is flanked by another DNA helitron AtREP3 in the opposite direction (Supplemental Figure 3), suggesting this region underwent rearrangements of TEs during evolution. The protein-coding sequence shows no similarity to any snRNA but contains a 3′-box-like sequence near the end of the imRNA reads (Figure 6; Supplemental Figure 3). Interestingly, expression of full-length SSP14 transcripts was specifically detected in CPL4RNAi seedlings but not in Col (Figure 6C), suggesting the production of SSP14 is regulated at the level of imRNA extension by the snRNA termination mechanism. The expression pattern of SSP14 in the wild-type Arabidopsis was strictly limited to pollens, where CPL4 expression was the lowest and the other Pol II-dependent snRNA loci showed 3′-extensions (Araport; https://apps.araport.org/thalemine/) (Loraine et al., 2013; Krishnakumar et al., 2015) (Supplemental Figure 4). Because the SSP14 and snR-DPG expression were similarly regulated by CPL4, hereafter, we collectively refer to both snR-DPG and SSP14 transcripts as “snR-DPG” transcripts, unless otherwise specified. The imRNA produced from SSP14 was termed imRNASSP14.
Figure 6.
SSP14 Locus Produces Unstable Short Transcripts in the Presence of Wild-Type CPL4, but Produces Stable Long Transcripts in CPL4RNAi.
(A) Top: imRNA-seq coverage for Col wild type and hen2-4 mutant. Bottom: RNA-seq coverage for Col and CPL4RNAi. An open triangle indicates the presence of 3′-box-like sequence. See Supplemental Figure 3 for the detailed features on the promoter and 5′-region of this locus.
(B) and (C) RT-PCR detection of 5′ short RNA (primer pair F472-F452) and long RNA (primer pair F472-F475). AT4G31430 is used as the control.
snR-DPG Transcripts Occur Widely in Other Plant Species
PIIsnRs are embedded in some transposons in plants, implying that snR-DPG-producing loci are present in diverse plant species in addition to Arabidopsis. BLAST searches using Arabidopsis snRNA sequences against the RefSeq mRNA database detected 150 potential snR-DPG transcripts from both dicots and monocots, and one species of fish (Supplemental Data Set 3). EST clones continuously spanning snRNA sequence and downstream protein-coding regions were found for 17 genes (Table 5), validating the snR-DPG expression from these loci. Combined with the observation that PIIsnR sequences embedded in TE in other green plants, these findings indicate that snR-DPG loci can frequently occur in plants. A few combinations of snR-DPG seem to be conserved, such as U12-GPI19 and U1-arsenate reductase, while others are unique to each plant species, suggesting neutral nature of the evolution of snR-DPG combinations.
Table 5. snRNA-DPG Transcripts Supported by ESTs Spanning snRNA-DPG in Various Organisms.
| Query | RefSeq | Species | snRNA-Extension EST | Downstream Protein-Coding Gene |
|---|---|---|---|---|
| AtU1 | NM_118471 | Arabidopsis thaliana | EG447525.1 | NAD(P)-binding Rossmann-fold superfamily protein |
| AtU5 | NM_115444 | Arabidopsis thaliana | EG419000.1 | LAF3-duplicate (upstream of LAF3, flanked by TE) |
| AtU12 | XM_013892844 | Brassica napus | ES911942.1 | GPI19 |
| AtU2 | XM_009128676 | Brassica rapa | DY013485.1 | F-box/FBD/LRR-repeat protein (AT5G56420-like) |
| AtU4 | XM_006450628 | Citrus clementina | DY288252.1 | Hypothetical protein |
| AtU1 | XM_008452530 | Cucumis melo | JG490974.1 | N-acetyltransferase p20-like |
| AtU1 | XM_008463125 | Cucumis melo | JG468085.1 | F-box/WD-40 repeat-containing protein (AT3G52030-like) |
| AtU2 | XM_008456053 | Cucumis melo | JG501029.1 | Cytochrome c oxidase subunit 5b-1 |
| AtU1 | XM_015757279 | Oryza sativa | CT861291.1 | Arsenate reductase 2.1 |
| AtU1 | XM_015777263 | Oryza sativa | AU173873.1 | d-xylose-proton symporter-like 3 |
| AtU2 | XM_015789462 | Oryza sativa | CI285625.1 | Proline-, glutamic acid-, and leucine-rich protein 1 |
| AtU2 | XM_015772229 | Oryza sativa | CB682952.1 | Synthesis of cytochrome oxidase (SCO1) homolog 1 |
| AtU2 | XM_015771499 | Oryza sativa | CI711653.1 | Zinc finger CCCH domain-containing protein 14 |
| AtU1 | NM_001329196 | Zea mays | FL163030.1 | Hypothetical protein |
| AtU4 | NM_001196788 | Zea mays | DV511687.1 | Cysteine synthase |
| AtU1 | XM_010236666/XM_003574239 | Brachypodium distachyon | DV482065.1 | Arsenate reductase 2.1 |
| AtU4 | NM_001292254 | Callorhinchus milii | JK858370.1 | Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform |
Potential snR-DPG transcripts bearing snRNA sequences on their 5′-end are retrieved from NCBI database by BLAST search (see Methods). Transcripts with corresponding ESTs are shown. See Methods for the search procedure.
Accumulation of snR-DPG Transcripts in CPL4RNAi Depends on SRD2
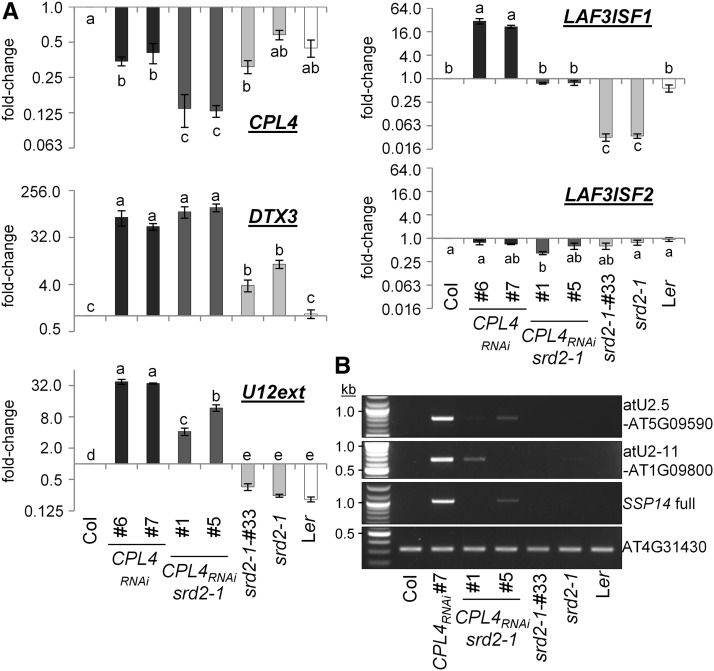
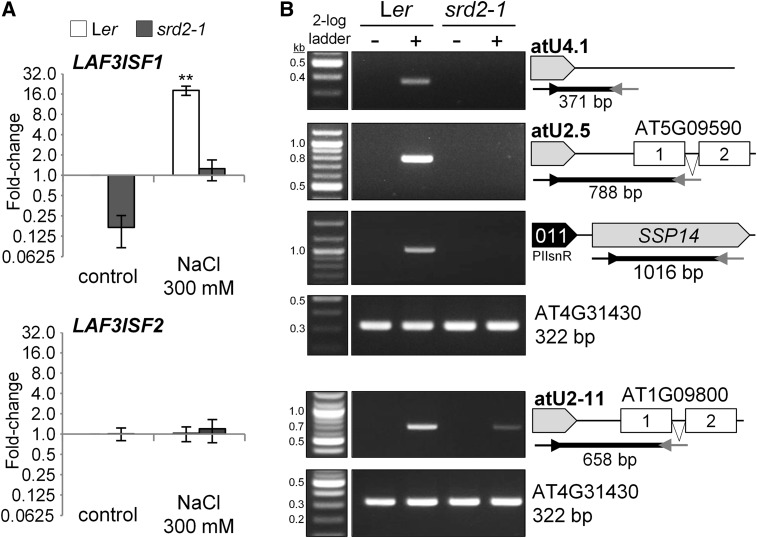
To establish that expression of snR-DPG loci is under the control of the snRNA transcription machinery, we analyzed the effect of the srd2-1 mutation on expression of several representative snR-DPG loci in the wild type and the CPL4RNAi background (Figure 7). The srd2-1 allele encodes a temperature-sensitive form of the SNAPc 50 subunit required for transcription activation of snRNA (Yasutani et al., 1994). The CPL4RNAi srd2-1 double homozygous seedlings were isolated after the genetic cross. The CPL4RNAi srd2-1 line exhibited a smaller cotyledon phenotype similar to the parental CPL4RNAi line and slightly shorter roots (Supplemental Figure 5). Before analysis, we confirmed that the CPL4RNAi srd2-1 maintained CPL4RNAi effects by testing the transcript levels of CPL4 and DTX3, as upregulation of DTX3 provides a marker for the effect of CPL4RNAi (Figure 7A; Supplemental Table 1; Fukudome et al., 2014). When we compared expression of LAF3 isoforms in srd2-1 mutant and parental wild type (Ler-0) under standard conditions (25°C), LAF3ISF1 accumulation was specifically decreased in srd2-1 while LAF3ISF2 was unaffected, confirming that LAF3ISF1 but not LAF3ISF2 is controlled by the snRNA transcription machinery (Figure 7A; Supplemental Table 1). Enhanced transcript levels of selected snR-DPG loci, as well as that of extended imRNASSP14, were decreased substantially in CPL4RNAi srd2-1 (Figures 7A and 7B). Therefore, we concluded that expression of snR-DPG in general is regulated by the snRNA transcription machinery.
Figure 7.
Accumulation of Polyadenylated snR-DPG Transcripts in CPL4RNAi Depends on snRNA-Activating Protein Complex Subunit SRD2.
(A) RT-qPCR analysis of CPL4RNAi x srd2-1 double homozygous lines (F4 generation) and respective parents (ecotypes; Col for CPL4RNAi [♀] and Ler for srd2-1 [♂]). srd2-1 #33 is a null segregant (srd2-1/srd2-1, without CPL4RNAi transgene) isolated from the same F2 population. Mean fold-change values relative to Col are plotted in log2 scale. Error bars indicate se of the mean of biological triplicates. Different letters indicate significant difference (one-way ANOVA followed by Tukey HSD test, P < 0.05).
(B) RT-PCR analysis of representative snR-DPG loci.
Salt Stress Triggers snRNA to snR-DPG Switching in the Wild Type in an SRD2-Dependent Manner
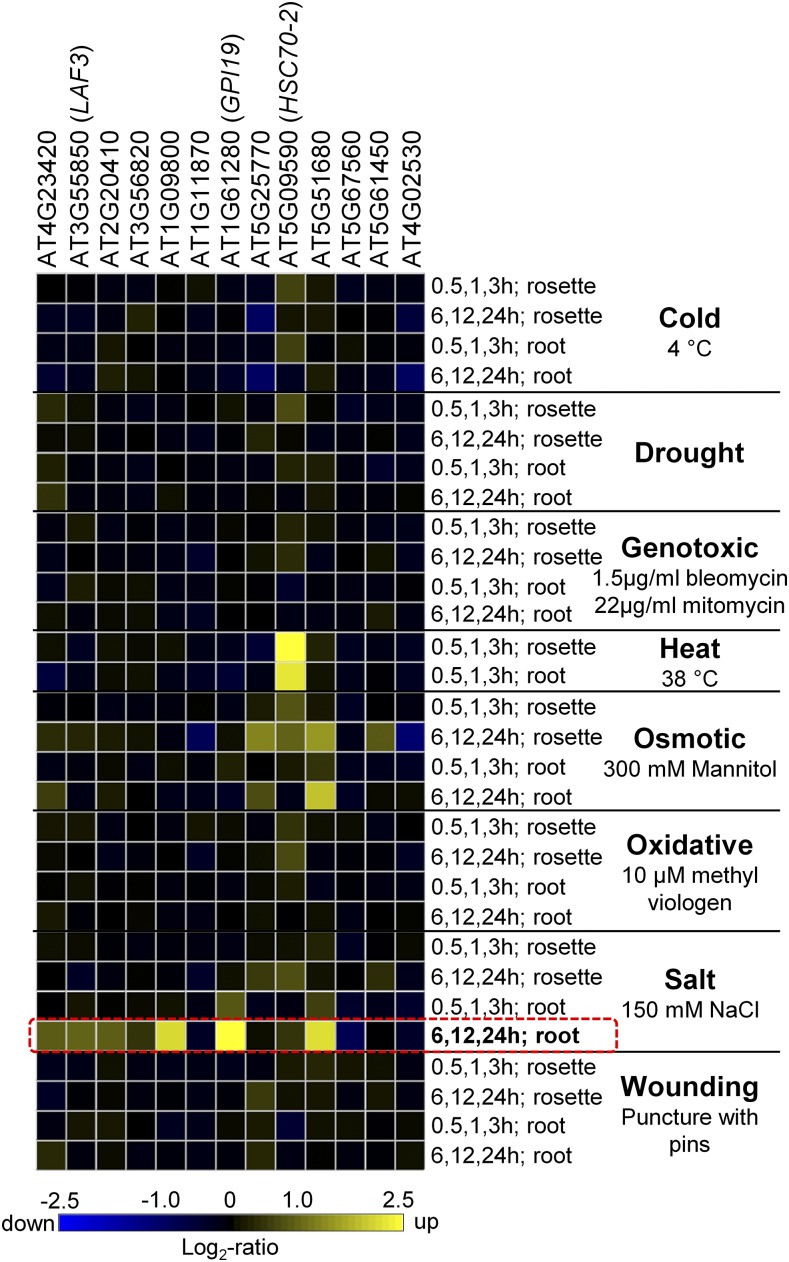
The genome-wide activation of snR-DPG transcription in CPL4RNAi indicates that Pol II CTD plays an active role in the expression of a specific subset of genes; however, little is known about the cues that trigger expression of snR-DPG. Based on a growing notion that the transcriptional responses to environmental stress involve phosphoregulation of the Pol II CTD (Lavoie et al., 1999; Fuda et al., 2012; Li et al., 2014), we hypothesized that snR-DPG expression could be activated by environmental stress. To test this hypothesis, we searched the AtGenExpress microarray expression data set for a condition that upregulates these DPGs. We found that salt treatment induces many of the DPGs that are located within 500 bp from an snRNA, especially in the root (Figure 8; Supplemental Figure 6). We also examined an RNA-seq experiment including salt-stressed samples (Cui et al., 2016) and recovered reads corresponding to snRNA-extended regions in the salt-stressed seedlings (Supplemental Figure 7).
Figure 8.
DPGs Near snRNA Are Upregulated in Roots Treated with Salt.
Relative expression levels of 13 DPG loci located within 500 nucleotides from Pol II-dependent snRNA from Table 2 (no ATH1 probe is available for AT1G16825) in response to various abiotic stresses. Data were retrieved from AtGenExpress microarrays (under Experiment AT-00120 in Genevestigator; 30 perturbations; 243 samples). For the salt (150 mM NaCl) treatments, “early” and “late” indicate “0.5, 1, 3 h” and “6, 12, 24 h,” respectively. DPGs are sorted by the distance from upstream snRNA (the closest is on the left).
To independently validate salt induction of snR-DPG and test if it is via the snRNA expression mechanism, we analyzed snR-DPG expression in salt-treated wild-type (Ler-0) and srd2-1 plants. Interestingly, salt treatment induced accumulation of snR-DPGs including LAF3ISF1 and SSP14 transcripts specifically in Ler-0 wild type but not in the srd2-1 mutant (Figure 9). The snRNA-independent LAF3ISF2 was not affected by the treatment. Importantly, atU4.1 snRNA locus without DPG also shows 3′-extended transcripts, indicating that the induction is independent of DPG-promoter (Figure 9). These results demonstrate that the snR-DPG accumulations are induced by salt stress through snRNA transcription and 3′-extension. Similar snR-DPG accumulation in response to salt was observed in Col seedlings as well, but CPL4RNAi did not show further induction of snR-DPG by the treatment (Supplemental Figure 8).
Figure 9.
Salt-Inducible Accumulation of snR-DPG Transcripts in the Wild Type Depends on SRD2.
RT-qPCR (A) and RT-PCR (B) analyses of total RNA prepared from the wild type (ecotype Ler) and srd2-1. The “+” or “−” indicate with or without salt treatment (300 mM NaCl for 200 min). Mean fold-change values relative to untreated Ler were plotted in log2 scale. Error bars show se of the mean for biological triplicates. **P < 0.01 by two-tailed Student’s t test between untreated Ler and each sample. AT4G31430 is used as the control.
Salt Stress Alters Pol II-CTD Phosphorylation Status and Pol II Occupancy on snR-DPG
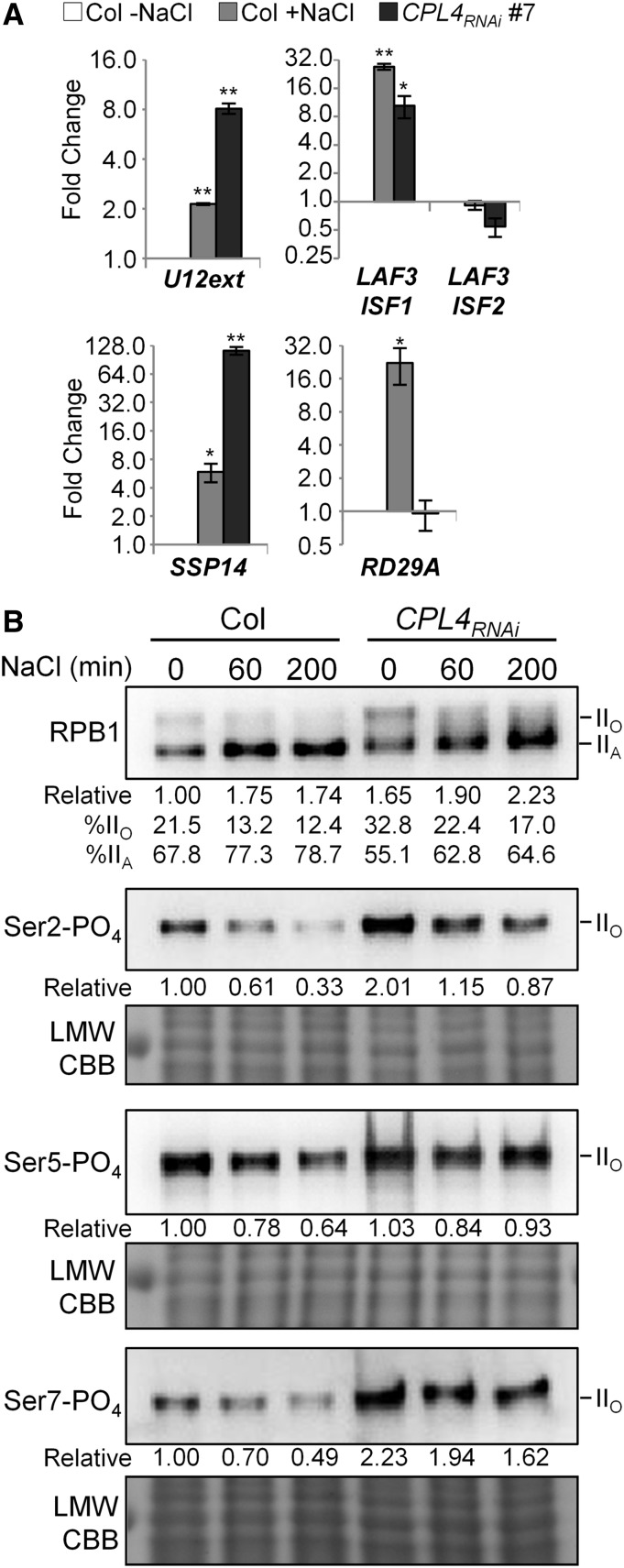
The similarity of snR-DPG expression patterns induced by salt treatment to that of CPL4RNAi lines suggested that salt stress may affect Pol II-CTD phosphorylation status. Therefore, we proceeded to characterize relationship among salt stress, Pol II-CTD phosphorylation, and snR-DPG expression. Due to the slow growth and reduced fertility of intact CPL4RNAi plants, we used an Arabidopsis callus system, which was established previously (Fukudome et al., 2014). The RT-qPCR analysis confirmed that the wild-type Col and CPL4RNAi calli reproduced snRNA 3′-extension phenotype of seedlings (Figure 10A). Immunoblot analysis of CTD phosphorylation profiles showed the reduced level of hyperphosphorylated Pol IIO form and accumulation of hypophosphorylated Pol IIA form in response to the salt treatment. This result indicated the overall Pol II-CTD phosphorylation levels were indeed reduced (Figure 10B). Analyses using position-specific anti-CTD-PO4 antibodies revealed that salt stress decreased Ser2-PO4/Ser5-PO4/Ser7-PO4 marks in the wild type (Figure 10B; Supplemental Figure 9). This was in contrast to the observation that CPL4RNAi showed a constitutively high level of CTD-PO4 marks, in particular, Ser2-PO4/Ser7-PO4 (Figure 10B) (Fukudome et al., 2014). Salt treatment also reduced the Pol II-CTD phosphorylation marks in CPL4RNAi; however, the level of CTD-PO4 in CPL4RNAi remained higher than that in the wild-type control even after the salt treatment.
Figure 10.
Salt Stress Causes Pol II-CTD Dephosphorylation.
(A) Relative expression levels of selected 3′-extended snRNAs in calli 2 d after transfer with or without salt treatment (300 mM, 200 min) by RT-qPCR. Mean fold-change values relative to untreated Col were plotted in log2 scale. Error bars show se of the mean for biological triplicates. *P < 0.05 and **P < 0.01 by two-tailed Student’s t test between untreated Col and each sample.
(B) Pol II-CTD phosphorylation levels detected by immunoblotting. Twelve to twenty-four micrograms of total proteins from calli 2 d after transfer treated with 300 mM NaCl for indicated time were analyzed. The antibodies used for the analysis re anti-RPB1 for total Pol II and phosphoserine-specific antibodies (3E10 for Ser2-PO4, H14 for Ser5-PO4, and 4E12 for Ser7-PO4). For each membrane, Coomassie blue (CBB) staining of low molecular weight area of the same gel not subjected to the blotting is shown as the loading control. Data taken from the same membrane after stripping and reprobing were stacked. Numbers show the relative band intensity compared with that of the wild type (Col). For RPB1, percentages of IIO and IIA form in total intensity observed were shown as well.
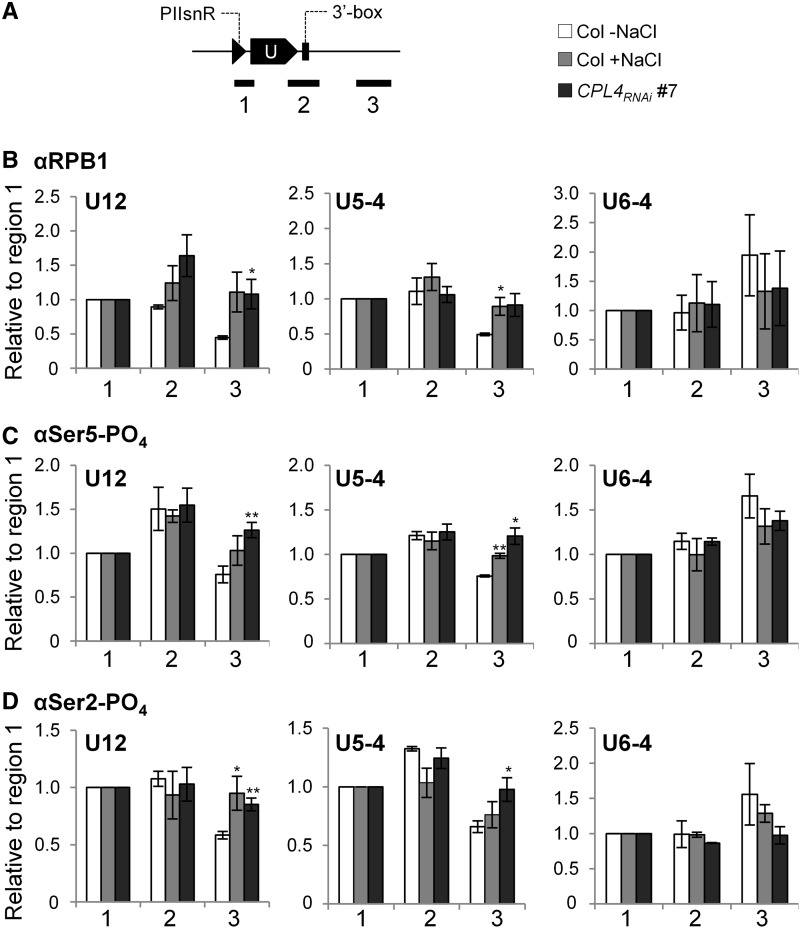
The immunoblot results were unexpected because salt treatment and CPL4RNAi had opposite effects on Pol II-CTD phosphorylation levels even though both result in similar snRNA extension and resulting snR-DPG accumulation in the host cells. Because immunoblots assess CTD phosphorylation status as a whole, but not at the level of individual genes, we performed chromatin immunoprecipitation (ChIP) assays to test if Pol II loading and CTD phosphorylation levels at the snRNA locus change during the salt-induced snRNA 3′-extension (Figure 11; Supplemental Figure 10). Of Pol II-dependent snRNA loci showing 3′-extension, U12 and U5-4 were analyzed because they were relatively distant from other genes that can interfere with the specific detection of the Pol II at the snRNA loci. A Pol III-dependent U6-4 was used for negative control. The ChIP qPCR regions selected for the analysis were a 5′ region including promoter and a part of mature snRNA (region 1), a region covering the 3′ end of snRNA and the 3′-box region (region 2), and a 3′ distal region downstream of the 3′-box (region 3) (Figure 11A).
Figure 11.
Pol II Occupancy on the Selected snRNA Loci in the Wild-Type Calli with or without Salt Treatment and in the Untreated CPL4RNAi Calli.
(A) Schematic diagram of ChIP target regions. Target regions for quantitative PCR analysis are indicated by black lines. Regions 1, 2, and 3 include PIIsnR (black triangle), mature snRNA (filled box arrow), 3′-box (filled box), and 3′-extended regions, respectively.
(B) to (D) The amount of Pol II on each target region were determined by ChIP-qPCR analysis using αRPB1 (B), αSer5-PO4 (C), and αSer2-PO4 (D) antibodies. Graphs show signal levels relative to the region 1. Error bars represent se of biological triplicates. *P < 0.05 and **P < 0.01 by one-tailed Student’s t test between untreated Col and each sample.
For U12 locus, loading of RPB1 (total Pol II) in the untreated Col cells decreased at region 3 relative to region 1, whereas CPL4RNAi produced a significantly higher level of RPB1 at region 3. The salt-treated Col cells also showed a higher level of RPB1 at region 3, but it was not statistically significant (P = 0.07, Student’s t test) (Figure 11B). The occupancy of Pol II with the Ser5-PO4 or the Ser2-PO4 marks showed different trends. The Ser5-PO4 ChIP profile peaked at the region 2 in all three samples. On the other hand, the Ser2-PO4 levels showed a similar trend to RPB1, with decreased signal levels in untreated Col cells at region 3, but significantly higher levels in salt-treated Col and CPL4RNAi cells. Analysis of U5-4 locus produced a similar trend but with a smaller magnitude, consistent with the only ∼20% expression level of the 3′-extended transcripts compared with the U12 locus. By contrast, ChIP signals observed at Pol III-dependent U6-4 locus did not show similarity to U12 or U5-4, suggesting Pol II association with U6-4 locus is not linked to Pol II transcriptional activity at the U6-4 locus. Overall, snRNA 3′-extension was commonly associated with increased Pol II marks in the downstream region of snRNA loci in CPL4RNAi and salt-treated wild-type cells.
DISCUSSION
Pol II-CTD phosphoregulation governs transcription of both protein-coding and noncoding genes. Although plant CTD kinases and CTD phosphatases involved in the regulation have been identified and characterized, little is known about the significance of the phosphoregulation in noncoding RNA transcription in plants. In this study, we show that a CTD phosphatase, CPL4, plays a pivotal role in choosing the fate of Pol II-transcribed U snRNA either for transcription termination/3′-processing or read-through transcription to produce 3′-extended snRNA. The data presented here support the model that the latter is not a mere transcriptional abnormality but is a part of transcriptional regulatory strategies in plants. Furthermore, we found that plants use imRNA as extension substrates to generate protein-coding mRNA. These snRNA/imRNA-to-mRNA switches occur not only in CPL4RNAi plants but have also been observed in the wild-type when plants are environmentally challenged (salt stress) and perhaps in specific tissues like pollen, and these switches are likely associated with alteration of CTD phosphorylation status. This indicates that snRNA/imRNA extension is a regulatory mechanism for plant gene expression and the Pol II CTD functions as a hub for this regulation.
Knockdown of CPL4 Causes snRNA 3′-Extension, Leading to Translatable snR-DPG Fusion
The regulation of snRNA transcription by Pol II-CTD phosphorylation has been characterized mostly in vertebrates, and little is known in plants; in particular, nothing is known about the termination signal for plant snRNA transcription. However, a recent study established that plants use the Integrator complex, the snRNA 3′-processing mechanism, similar to animals. In animals, Ser2-PO4/Ser7-PO4 double phosphorylation marks facilitate snRNA 3′-end processing, whereas Ser5 phosphorylations were inhibited by differentially impacting recruitment of the Integrator complex to Pol II transcribing snRNA (Egloff et al., 2010). Based on the retention of Pol II marks at the downstream (region 3) in CPL4RNAi and salt-treated wild-type Col cells, we can speculate that the high CTD-PO4 marks, such as Ser5-PO4, at the 3′ region direct Pol II to continue transcription elongation, instead of transcription termination and recruitment of the Integrator complex.
This implies that Ser5-PO4 phosphatase activity of CPL4 (Fukudome et al., 2014) may play an active role in snRNA termination in Arabidopsis. In animals, two conserved CTD phosphatases involved in Ser5 dephosphorylation during snRNA transcription termination have been reported. RNA Pol II-associated protein 2 (RPAP2) is recruited by Ser7-PO4 and specifically dephosphorylates Ser5-PO4, which in turn recruits the Integrator complex (Egloff, 2012; Egloff et al., 2012). Another CTD phosphatase, SSU72, which dephosphorylates Ser5-PO4 and Ser7-PO4, is required for proper snRNA 3′-end processing in both animals and yeast (Wani et al., 2014). Surprisingly, there has been no implication of functional association of FCP1 (TFIIF-interacting CTD Phosphatase1)-family CTD phosphatases in the snRNA transcription cycle. Therefore, our data suggest an additional function for FCP1-family CTD phosphatase in snRNA transcription and 3′-end processing, at least in plants. Because both RPAP2 and SSU72 homologs are also encoded by the Arabidopsis genome, it is not clear whether CPL4 replaces all or a part of the RPAP2/SSU72 function in plants or acts in concert with these phosphatases.
Transposon-Embedded PIIsnR
Although snRNAs and their transcriptional mechanisms are conserved in eukaryotes, the plant genome structures uniquely connect snRNA 3′-extension with the production of snR-DPG transcripts. In the Arabidopsis genome, 75 snRNA genes are evenly distributed on the chromosomes, with several small gene clusters (Kaul et al., 2000; Wang and Brendel, 2004). Thirty-three Pol II-dependent snRNA genes are flanked by protein-coding genes that are located within 1 kb downstream of the snRNA loci; 18 and 15 of them are on the same and the opposite strand, respectively (Table 2). In contrast, human snRNA loci are flanked with long intergenic regions without protein-coding potential. All functional human U1 genes have ∼2-kb conserved flanking sequences in both upstream and downstream regions (Manser and Gesteland, 1982; Htun et al., 1984), many of which extend to 20 to 24 kb extensively conserved intergenic sequences for both directions (Bernstein et al., 1985). Human U2 snRNA genes are clustered as 10 to 20 tandem repeats of a 6-kb unit (Vanarsdell and Weiner, 1984; Westin et al., 1984; Lindgren et al., 1985). Another plant genome feature that facilitates the integration of snRNA transcription mechanisms into a unique gene expression system is the PIIsnR sequence in DNA transposons, which provides mobilization and propagation potentials to PIIsnRs. In the Arabidopsis genome, 75 PIIsnRs are located inside of nonautonomous DNA transposons, particularly in the ATREP5 subclass. These transposon-born promoters are indistinguishable from functional snRNA gene promoters. This may represent an ancient gene capture event(s) by ATREP5, which performs rolling-circle transposition and can incorporate and spread the flanking genomic sequences (Kapitonov and Jurka, 2001, 2007). During genome evolution, the snRNA promoters mobilized by transposons could be a source of de novo gene expression, mediated by Pol II-CTD phosphoregulation. Indeed, the novel imRNA we identified (imRNASSP14) is expressed from PIIsnR associated with an ATREP fragment. Although some characteristics of imRNASSP14 resemble previously reported ncRNAs in animals and fungi (Wyers et al., 2005; Kapranov et al., 2007), its production from a PIIsnR, instability, and potential to extend to produce mRNA make imRNASSP14 distinct. Based on CPL4-regulated conversion to mRNA, transcription termination of imRNASSP14 is likely under the control of 3′-box sequence embedded in the SSP14 coding region. Because the 3′-box motif is relatively tolerant to deviation from the consensus sequence (Connelly and Filipowicz, 1993), the 3′-box-like sequences frequently occur in the Arabidopsis genome sequence, increasing the probability of PIIsnR-3′-box pairings during the genome evolution. imRNASSP14 also shares several features with upstream noncoding transcripts, ncRNAs that overlap with the 5′ region of pre-mRNAs and are degraded by the nuclear exosome pathway (Chekanova et al., 2007). Although a 3′-extension of the upstream noncoding transcript has not been studied, it is possible that conditional conversion of short-lived ncRNA to mRNA by 3′-extension occurs with other types of ncRNAs to regulate gene expression.
Salt Stress Induces Pol II-CTD Dephosphorylation and snR-DPG Accumulation
The most likely input point where salt signal fine-tunes the switch between snRNA termination and 3′-extension is the CTD phosphorylation status. Salt treatment caused global dephosphorylation of Pol II CTD (Figure 10). Signal-triggered Pol II-CTD phosphorylation/dephosphorylation are important for several developmental and stress-regulated gene expression systems in animals (Bellier et al., 1997; Dubois and Bensaude, 1998; Shim et al., 2002; Walker et al., 2007) and have also been proposed in plant innate immunity responses (Li et al., 2014). In the latter case, signals generated from a perception of pathogen-associated molecular patterns are transduced through a MAP kinase cascade and induce rapid and transient cyclin-dependent kinase C-mediated phosphorylation of Pol II CTD in Arabidopsis. By contrast, abiotic stress signal-induced CTD dephosphorylation of plant Pol II has not been reported, and stress-specific CTD phosphatase has not been identified, except by implication from our previous genetic studies (Koiwa et al., 2002; Xiong et al., 2002). Although our results demonstrated reduction of CTD phosphorylation after salt stress (Figure 10), the identity of the CTD phosphatase responsible for the salt-induced CTD dephosphorylation has not been determined unambiguously. CPL4, a major CPL in Arabidopsis, is a candidate for the salt-activated CTD phosphatase activity. However, the salt-induced decrease in the CTD phosphorylation level was also observed in CPL4RNAi. This could be due to the residual CPL4 activity in CPL4RNAi, but activation of other >20 CTD-phosphatase isoforms/homologs is possible (Koiwa, 2006).
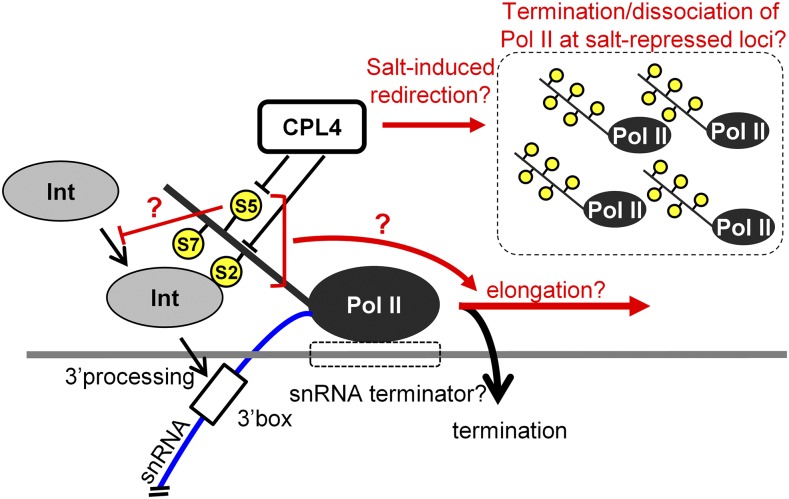
Despite the opposite direction of changes in Pol II-CTD phosphorylation profile caused by salt treatment and CPL4RNAi, CTD phosphorylation profiles at Pol II-dependent U12 and U5-4 snRNA loci were similar between these conditions. Both salt treatment and CPL4RNAi promoted retention of Ser2/Ser5 phosphorylation marks in U12 and U5-4 snRNA, even though the effect of salt was not as strong as CPL4RNAi. The retention of CTD-PO4 marks midst of overall CTD dephosphorylation during the salt stress suggests that the stress-induced CTD dephosphorylation has specificity toward Pol II not engaged in snRNA transcription (Figure 12). In such a scenario, salt stress may redirect more CTD phosphatase activities from Pol II at snRNA loci to Pol II terminated at or dissociated from the salt-repressed loci to regenerate initiation-competent, free Pol II. This is consistent with the result reported for S. cerevisiae and D. melanogaster FCP1 showing that FCP1 prefers free Pol II over elongating Pol II (Kong et al., 2005; Fuda et al., 2012). This may promote retention of CTD-PO4 levels in elongating Pol II and continuation of transcription elongation at snRNA loci under the salt stress. Regulation of CTD phosphorylation is a complicated process with multiple factors providing inputs directly or indirectly, and more work needs to be done to understand the activities of these factors during the plant stress response.
Figure 12.
A Model for the Transcriptional Switching by Salt Stress and CPL4RNAi.
Black and red arrows indicate events during normal conditions and under salt stress, respectively. Yellow circles indicate serine phosphorylation at the Pol II CTD. Only a single CTD repeat is shown for the illustration. The blue line indicates an snRNA transcript. Under normal conditions, CPL4 functions in dephosphorylation of CTD-PO4 (possibly Ser5-PO4) of Pol II and promotes the snRNA transcription termination and the recruitment of the Integrator complex (Int). Gene repression events associated with salt stress increase the level of termination/dissociation of Pol II complexes in the cell; these free complexes recruit CPL4 and other CTD phosphatases to regenerate pools of initiation-competent Pol II. Low availability of CPL4 activities at snRNA loci allows the local CTD phosphorylation levels to remain unchanged and promotes transcription elongation while preventing transcription termination and 3′-end processing. CPL4RNAi mimics low CPL4 availability by directly decreasing cellular CPL4 levels.
Plants Express Diverse SnR-DPGs
Adopting snRNA 3′-extension to their genome structure, plants produced a unique stress-inducible gene expression system, which is likely activated cotranscriptionally via alteration of Pol II-CTD phosphorylation level. This system resembles heat shock activation of paused Pol II in animals (Ni et al., 2004) and may allow a quicker response than de novo activation of promoters by transcriptional factors. The BLAST survey identified potential 150 snR-DPGs in Arabidopsis and other plant species, including 17 EST-supported snR-DPGs (Table 5; Supplemental Data Set 3), suggesting the snRNA-to-mRNA conversion is a ubiquitous process in plants. There are diversities in snR-DPG pairs in different plant genomes, but some conserved combinations, such as U12-GPI19 (Arabidopsis and Brassica napus) and U1-arsenate reductase 2.1 (O. sativa and Brachypodium distachyon), may have biological functions that were selected for during evolution (Table 5). However, the role of a specific combination needs to be empirically examined for their functions because it is not immediately obvious if snR-DPG-encoded proteins collectively perform protective functions during the stress. Function through other mechanisms without protein production, for example, providing a physical scaffold (Vilborg et al., 2015) or production of regulatory RNAs (Uesaka et al., 2014; Vera and Dowell, 2016), cannot be excluded. Furthermore, the presence of a snRNA structure on an mRNA may confer new functionality to the mRNA, like the high systemic mobility of dicistronic mRNA-tRNA in Arabidopsis (Zhang et al., 2016). CPL4RNAi revealed hidden RNA dynamics orchestrated around plant Pol II CTD, which warrants further investigation.
METHODS
Plant Materials and Growth Conditions
Unless otherwise stated, seeds of CPL4RNAi lines previously established in Arabidopsis thaliana ecotype Columbia gl1 (Col) were sown on germination media (0.25× Murashige and Skoog [MS] salts, 0.5% sucrose, and 1.5% agar, pH 5.6 to 5.8) and were stratified at 4°C for 2 d (Bang et al., 2006). Then, they were grown vertically at 25°C under long-day conditions (16-h day using 50% output of Philips F17T8/TLT41 bulb/8-h night). Ten- to fifteen-day-old seedlings were used for experiments. srd2-1 in Landsberg erecta (Ler) background was kindly provided by Misato Ohtani and Munetaka Sugiyama. Callus cultures were induced from seedling explants placed on callus induction medium containing 1× MS salts, 2% sucrose, 0.2 g/L KH2PO4, 0.1 g/L myo-inositol, 1× B5 vitamins, 2.0 mg/L 2,4-D, 0.05 mg/L kinetin, and 0.9% agar. The callus cultures were maintained in dark and 25°C, and transferred to fresh callus induction medium once in every 7 to 10 d.
RT-PCR and qPCR
RNA extraction and reverse transcription were done as previously described (Fukudome et al., 2014). Seedlings or calli samples were separately salt-treated and/or harvested in triplicates. Total RNAs were extracted using TRIzol reagent, followed by a DNase I treatment to eliminate DNA contamination. One to two micrograms of total RNA was converted to the first-strand cDNA using random hexamers and GoScript Reverse Transcriptase (Promega) unless otherwise stated. Gene-specific primer cocktail (Supplemental Data Set 4) and an oligo(dT) primer were used for unstable short imRNASSP14 (Figure 6B) and polyadenylated snR-DPG transcripts (Figure 7), respectively. The reverse transcription products were analyzed using a LightCycler 480 (Roche Diagnostics) and Bullseye EvaGreen qPCR MasterMix (Midwest Scientific). GAPDH (AT1G13440) was used as an internal control for normalization in qPCR. In RT-PCR experiments of snR-DPGs, KAKU4 (AT4G31430) was chosen as a control due to its low expression level closer to these snR-DPGs and consistency of its expression in various conditions. Primers used are listed in Supplemental Data Set 4.
snRNA Detection by RNA Gel Blot
RNA gel blot analysis was conducted as described previously (Zhang et al., 2017). Probe sequences used are listed in Supplemental Data Set 4.
RNA-Seq
Ten-day-old Col wild-type and CPL4RNAi #7 seedlings horizontally grown on germination media were subjected to total RNA extraction using RNeasy Plant Mini kit (Qiagen). The total RNA was then submitted to Otogenetics for RNA-seq analysis (paired-end, 100 bp). After validating the integrity and purity of total RNA were using an Agilent Bioanalyzer and OD260/280, 2 µg of total RNA was used for rRNA depletion using the Ribo-Zero Magnetic Gold Kit (EpiCentre). Recovered total RNA from rRNA depletion was subjected to cDNA synthesis using SMARTer PCR cDNA synthesis kit (Clontech Laboratories; catalog no. 634925). Sequencing was performed on the Illumina HiSeq 2000 (Illumina) with chemistry v3.0 and using the 2×100-bp paired-end read mode and original chemistry from Illumina according to the manufacturer’s instructions.
For RNA-seq analysis of imRNAs, total RNAs were purified from 7-d-old Col-0 wild-type and hen2-4 mutant roots (Lange et al., 2014) growing on germination media using miRNA-easy kit (Qiagen). Sequencing libraries were prepared using TruSeq Stranded Total RNA with Ribo-Zero Plant kit. To enrich imRNA, fragmentation step was omitted and library cDNA with 100 to 500 nucleotides were purified using Pippin (Sage Science) before sequencing by Illumina HiSeq (125 nucleotides, paired-end).
Bioinformatics
For RNA-seq analysis, raw read files (fastq) from our experiments and the Sequence Read Archive were uploaded to Galaxy (https://usegalaxy.org/). All data analyses were performed on the Galaxy platform. After QC (FastQC) and adapter-trimming (Trim Galore! v0.4.0), the processed reads were mapped with Tophat (v2.0.14) against reference assembly Ensembl TAIR10 from Illumina iGenome (Supplemental Table 2). Because the original coordinates for each of 74 snRNAs from the ASRG database (Wang and Brendel, 2004) were not consistent with TAIR10 genome, we manually updated each coordinate by running a BLAST search (see Supplemental Data Set 1 for the updated coordinates for all snRNAs tested in this study). Coverage depth of each snRNA-extension region was calculated by SAMtools_BedCov (v1.2) and normalized by a total number of mapped reads (DCPM). Fragments per kilobase million values for LAF3 isoforms and fold-change values of DPGs were obtained by running Cuffdiff (v2.2.1.3). For coverage visualization in trackster, the mapping files (bam) were converted to bigwig format through BEDTools, Genome Coverage (v0.1.0), and Wig/BedGraph-to-bigWig converter (v1.1.0). The coverage was normalized by total number of mapped reads.
Pol II-Dependent snRNA Promoter Search
The Pol II-dependent promoter (PIIsnR) motifs (RTCCCACATCGN32-35TATAA) were searched against Arabidopsis TAIR10 genome sequence (Bao et al., 2015) using the Find Individual Motif Occurrences (FIMO) tool in the MEME suite (Bailey et al., 2009; Grant et al., 2011). The same search was performed against the consensus sequences of transposable elements in green plants in RepBase (Bao et al., 2015). Due to a size limitation of the input sequences, the search was separately conducted on chromosome 1-3 and chromosome 4-5 in the Arabidopsis genome search (parameters; match P value <1 × 10−6; scanning both strands). Total 150 matched sequences with q-value < 0.1 (corrected P value for multiple testing) were considered for further analysis. For each PIIsnR, coverage (DCPM) of 200-bp region in starting from 23 bp downstream of the TATA-box was computed as described above. The 150 sequences were classified into three classes (“Repeat,” “snRNA,” and “novel”) based on the phylogram computed and visualized by the multiple sequence alignment tool Clustal Omega and the TreeView software, respectively (Page, 1996; Sievers et al., 2011). For the search against TE consensus sequences, 11 matched sequences with q-value < 0.01 are listed in Table 3.
snR-DPG Search in Other Organisms
For identification of potential snR-DPG transcripts in other species, BLASTN search against Refseq_rna database using Arabidopsis U1, U2, U4, U5, and U12 snRNA sequence as query was performed. Transcripts in NM/XM (mRNAs, predicted model included) categories with similarity to the query snRNA sequences on the same strand were selected. Then, transcripts with “hit-start” position larger than the query snRNA length, and those with the alignment coverage <50% were filtered to focus on transcripts harboring snRNA sequences on 5′-end. In total, 150 Refseq_rna transcripts were obtained (Supplemental Data Set 3). Each of the 150 transcripts was then subjected to BLASTN search against the EST data set to find if there were supporting ESTs for the snRNA-DPG fusion transcript (Table 5).
U12-LUC Reporter System
A luciferase expression vector was prepared based on pEnEOiLUCThsp (GenBank accession no. KF545094.1) after replacing the LUC coding sequence with LUC2 (Promega). The wild-type U12-LUC construct was prepared using a 1.5-kb genomic fragment (chromosome 1, 22,602,141–22,603,607) corresponding between 1.0 kb upstream of the U12 transcription start site and 24 bp downstream of GPI19 start codon (ATG), which was placed upstream of LUC to generate translational fusion of GPI19-LUC driven by U12/GPI19 promoter. The m, 3m, and m3m variants were prepared by introducing mutations at USE (chromosome 1, 22,603,035–22,603,046, GTCCCACATCG to GgCaaACATCG) and/or 3′-box (chromosome 1, 22,603,073–22,603,079, AGTAAAT to TCGCGAC) by overlap extension protocol (Ho et al., 1989). See Supplemental Data Set 4 for primer sequences used. Resulting U12-LUC plasmids were recombined with pCB302-GW (a Gateway derivative of pCB302; Xiang et al., 1999) using LR clonase (Life Technologies). Resulting pCB302U12-LUC plasmids were introduced into Agrobacterium tumefaciens GV3101 and used for floral transformation of Col-0 wild type (Chung et al., 2000). The T1 transformants were selected on soil by spraying 30 µg/mL Liberty herbicide as described previously (Bang et al., 2006).
For luciferase assay, seeds of reporter transgenic plants were germinated on germination media supplemented with 10 µg/mL phosphinothricin. Seeds sown on media plates were stratified for 2 d and then kept at 25°C under a 16-h-light/8-h-dark cycle for 8 d. Luciferase activity was measured after spraying with 1 mM luciferin. Image acquisition with a CCD system and processing with Winview software (Roper Scientific) were performed as described previously (Koiwa et al., 2002).
Protein Extraction for RNA Pol II-CTD Phosphorylation
Callus tissues with or without salt treatment were homogenized in protein extraction buffer (50 mM Tris-HCl, pH 9.0, 100 mM NaCl, 12.5% glycerol, 2.5 mM EDTA, 10 mM β-mercaptoethanol, and 20 mM sodium fluoride) supplemented with 1 mM PMSF and 1× Proteinase inhibitor cocktail for plant cell and tissue extracts (Sigma-Aldrich). The crude extracts were then filtered through glass beads column to remove debris. The filtrate as total protein fraction was separated on 5% SDS-PAGE gel and subjected to immunoblot analysis.
Salt Treatment
Ten- to thirteen-day-old seedlings were placed onto two-layer of filter papers saturated with a solution containing 0.25× MS salts and 300 mM NaCl in a Petri dish for 30 to 200 min at 25°C with keeping a lid on. After treatments seedlings were snap frozen in liquid nitrogen and stored at −80°C until total RNA extraction described above. For salt treatment of calli, see below.
Pol II Occupancy Assessment by ChIP-qPCR
All ChIP experiments were conducted in triplicate biological replicates using cells that were salt-treated and harvested separately. Calli at 2 d after transfer were used. For salt treatment, wild-type (Col) calli were immersed in liquid callus induction medium containing 300 mM NaCl and incubated with gentle horizontal shaking at room temperature for 200 min. The container was covered with foil to block light. After salt treatment, NaCl-treated calli were harvested along with untreated wild-type and CPL4RNAi calli. Subsequently, ChIP was conducted as previously described, with some modifications (Saleh et al., 2008; Castillo-González et al., 2015).
The harvested calli were subjected to cross-linking using vacuum infiltration system. Calli were immersed in cross-linking buffer containing 0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1% formaldehyde, and 1 mM PMSF and then were vacuum infiltrated for 15 min (2 min vacuum, release, 8 min vacuum, release, and additional 5 min vacuum). The formaldehyde was quenched by adding final 100 mM glycine to the solution, followed by additional 5 min vacuum infiltration. The cross-linked materials were washed five times with ice-cold water, then flash frozen in liquid nitrogen. The frozen cells were ground into powder, and resuspended in 5 volumes of ice-cold HONDA buffer (0.44 M sucrose, 1.25% Ficoll, 2.5% Dextran T40, 20 mM HEPES, pH7.4, 10 mM MgCl2, 0.5% Triton X-100, 5 mM DTT, 1 mM PMSF, and 1× Protease inhibitor cocktail for plant cell and tissue extracts [Sigma-Aldrich; P9599]). The homogenized slurry was filtered through two layers of Miracloth (EMD Millipore). The filtrates were centrifuged at 2000g for 15 min at 4°C. After removing supernatant, the pellet was resuspended in 500 μL to 1 mL Nuclear Lysis buffer containing 50 mM Tris-HCl, pH 8.0, 10 mM EDTA, pH 8.0, 1% SDS, 1 mM PMSF, and 1× Protease Inhibitor Cocktail. Chromatin-DNA was sheared into ∼500-bp fragments on ice using a Sonic Dismembrator 60 (Fisher Scientific), 10 cycles of 15-s pulse with a 1-min interval between each cycle to prevent overheating. The sonicated solution was centrifuged at 15,000 rpm for 10 min at 4°C to precipitate debris. The sonicated chromatin solution was then aliquoted and stored in −80°C. For immunoprecipitation, the sonicated chromatin containing 20 to 30 µg DNA was diluted 10-fold with Chromatin Dilution buffer (16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1.2 mM EDTA, and 1.1% Triton X) to a final volume of 500 μL in a 1.5-mL nonstick tube. Then, the diluted chromatin solution was incubated with antibodies for overnight at 4°C. Amount of antibody used were 8.64 µg for anti-RPB1 polyclonal antibody and 5 µg for anti-Ser2-PO4 (Abcam ab5095) and anti-Ser5-PO4 polyclonal antibody (Abcam ab5131). Following the overnight incubation of antibody-sonicated chromatin mixture, 30 to 40 μL of Dynabeads Protein G (Thermo Fisher) washed with Binding/Washing buffer (BW buffer; 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS, and 1 mM PMSF) were added to the mixture. The mixture was further incubated for 2.5 h at 4°C with gentle rotation. Then, the beads were magnetically separated from the mixture and washed three times with 1 mL BW buffer, followed by 2 washes with 1 mL TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA). Each wash was 5 min of rotation at 4°C. After the final wash, beads were transferred to a 0.6-mL tube and the supernatant was completely removed. Subsequent reverse cross-link and elution procedures follow the Chelex-based method previously described (Nelson et al., 2006). To the washed beads, 50 μL of 10% Chelex (w/v, in water) solution was added. Also, 0.5 μL of the original sonicated chromatin was added to 49.5 μL of 10% Chelex to prepare 1% input DNA solution. Chromatin-DNA complexes were reverse cross-linked by incubating the beads-Chelex solution at 100°C for 10 min. Then, 0.5 μL of protease K (Invitrogen) was added to each tube and incubated at 43°C for 1 h. After the protease K treatment, the enzymes were deactivated by incubating the tube at 100°C for 10 min. The supernatant (∼35 μL recoverable without taking Chelex bed) containing eluted DNA was analyzed by quantitative PCR as described above. No antibody was added to negative control samples (NoAb).
Accession Numbers
Nucleotide sequence data from this article can be found in the EMBL/GenBank data libraries under Arabidopsis AGI locus identifiers AT1G61280 (GPI19), AT1G20320 (SSP14), AT3G55850 (LAF3), AT1G28560 (SRD2), AT5G58003 (CPL4), AT2G04050 (DTX3), AT4G31430 (KAKU4), and AT1G13440 (GAPDH). RNA-seq data used in this study are deposited in Gene Expression Omnibus database under accession number GSE98047.
Supplemental Data
Supplemental Figure 1. Detection of 3′-extended snRNA transcripts in CPL4RNAi line by RT-PCR.
Supplemental Figure 2. Classification of Arabidopsis Pol II-dependent snRNA promoter motifs.
Supplemental Figure 3. Features overlapping with the Pol II-dependent snRNA promoter region SSP14.
Supplemental Figure 4. SSP14 expression and snRNA 3′-extensions in pollen.
Supplemental Figure 5. The growth of CPL4RNAi srd2-1 plants
Supplemental Figure 6. Extended Figure 8, including expression levels in other AtGenExpress microarray sets including biotic stress and hormone treatments.
Supplemental Figure 7. Detection of snRNA-3′ extension in a salt RNA-seq study.
Supplemental Figure 8. Salt-inducible accumulation of 3′-extend end snR-DPG transcripts.
Supplemental Figure 9. Salt-induced Pol II-CTD dephosphorylation detected by polyclonal antibodies.
Supplemental Figure 10. Summary graphs of %input values from ChIP-qPCR experiments used to determine relative Pol II occupancy at snRNA 3′-extended regions.
Supplemental Table 1. ANOVA tables for Figures 1F and 7A.
Supplemental Table 2. Numbers of mapped reads in the RNA-seq experiments.
Supplemental Data Set 1. Detection of snRNA and snRNA 3′-extension in CPL4RNAi RNA-seq.
Supplemental Data Set 2. List of 150 PIIsnRs and associated downstream expression levels (DCPM) in imRNA-seq.
Supplemental Data Set 3. List of 150 snR-DPG transcripts found in RefSeq database.
Supplemental Data Set 4. List of primers/probes used in this study.
Acknowledgments
We thank Charles D. Johnson and Richard Metz (Texas A&M AgriLife Genomics and Bioinformatics Service) for his support on the im-ncRNA-seq experiment. We also thank Misato Ohtani (NARA Institute of Science and Technology) and Munetaka Sugiyama (University of Tokyo) for kindly providing srd2-1 seeds. This work was supported by the National Science Foundation (MCB0950459 and MCB1253369), USDA-CSREES (2010-34402-20875) “Designing Food for Health,” Texas AgriLife Research Genomics of Plant Water Use Seed Grant. A.F. was supported by the Student Exchange Support Program (scholarship for long-term study abroad from Japan Student Services Organization) and Dissertation Fellowship from Office of Graduate and Professional Studies, Texas A&M University. D.S. was supported by a fellowship from the China Scholar Council.
AUTHOR CONTRIBUTIONS
H.K. supervised the project. A.F., D.S., X.Z., and H.K. designed and performed the experiments. A.F., D.S., X.Z., and H.K. analyzed the data. A.F., X.Z., and H.K. wrote the article.
References
- Allison L.A., Wong J.K.C., Fitzpatrick V.D., Moyle M., Ingles C.J. (1988). The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol. Cell. Biol. 8: 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang W., Kim S., Ueda A., Vikram M., Yun D., Bressan R.A., Hasegawa P.M., Bahk J., Koiwa H. (2006). Arabidopsis carboxyl-terminal domain phosphatase-like isoforms share common catalytic and interaction domains but have distinct in planta functions. Plant Physiol. 142: 586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W., Kojima K.K., Kohany O. (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellier S., Dubois M.F., Nishida E., Almouzni G., Bensaude O. (1997). Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol. Cell. Biol. 17: 1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein L.B., Manser T., Weiner A.M. (1985). Human U1 small nuclear RNA genes: extensive conservation of flanking sequences suggests cycles of gene amplification and transposition. Mol. Cell. Biol. 5: 2159–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet F., Vigneron M., Bensaude O., Dubois M.F. (1999). Transcription-independent phosphorylation of the RNA polymerase II C-terminal domain (CTD) involves ERK kinases (MEK1/2). Nucleic Acids Res. 27: 4399–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.J., Hendrich B.D., Rupert J.L., Lafrenière R.G., Xing Y., Lawrence J., Willard H.F. (1992). The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542. [DOI] [PubMed] [Google Scholar]
- Castillo-González C., Liu X., Huang C., Zhao C., Ma Z., Hu T., Sun F., Zhou Y., Zhou X., Wang X.J., Zhang X. (2015). Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. eLife 4: e06671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao S.H., Greenleaf A.L., Price D.H. (2001). Juglone, an inhibitor of the peptidyl-prolyl isomerase Pin1, also directly blocks transcription. Nucleic Acids Res. 29: 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova J.A., et al. (2007). Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353. [DOI] [PubMed] [Google Scholar]
- Chung M.-H., Chen M.-K., Pan S.-M. (2000). Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res. 9: 471–476. [DOI] [PubMed] [Google Scholar]
- Connelly S., Filipowicz W. (1993). Activity of chimeric U small nuclear RNA (snRNA)/mRNA genes in transfected protoplasts of Nicotiana plumbaginifolia: U snRNA 3′-end formation and transcription initiation can occur independently in plants. Mol. Cell. Biol. 13: 6403–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P., Chen T., Qin T., Ding F., Wang Z., Chen H., Xiong L. (2016). The RNA polymerase II C-terminal domain phosphatase-like protein FIERY2/CPL1 interacts with eIF4AIII and is essential for nonsense-mediated mRNA decay in Arabidopsis. Plant Cell 28: 770–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L., Muniz L., West S. (2014). 3′ End formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 28: 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M.F., Bensaude O. (1998). Phosphorylation of RNA polymerase II C-terminal domain (CTD): a new control for heat shock gene expression? Cell Stress Chaperones 3: 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M.F., Bellier S., Seo S.J., Bensaude O. (1994). Phosphorylation of the RNA polymerase II largest subunit during heat shock and inhibition of transcription in HeLa cells. J. Cell. Physiol. 158: 417–426. [DOI] [PubMed] [Google Scholar]
- Egloff S. (2012). Role of Ser7 phosphorylation of the CTD during transcription of snRNA genes. RNA Biol. 9: 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., Zaborowska J., Laitem C., Kiss T., Murphy S. (2012). Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol. Cell 45: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., Szczepaniak S.A., Dienstbier M., Taylor A., Knight S., Murphy S. (2010). The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J. Biol. Chem. 285: 20564–20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., O’Reilly D., Chapman R.D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. (2007). Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda N.J., Buckley M.S., Wei W., Core L.J., Waters C.T., Reinberg D., Lis J.T. (2012). Fcp1 dephosphorylation of the RNA polymerase II C-terminal domain is required for efficient transcription of heat shock genes. Mol. Cell. Biol. 32: 3428–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome A., Aksoy E., Wu X., Kumar K., Jeong I.S., May K., Russell W.K., Koiwa H. (2014). Arabidopsis CPL4 is an essential C-terminal domain phosphatase that suppresses xenobiotic stress responses. Plant J. 80: 27–39. [DOI] [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., Noble W.S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B., Eick D., Bensaude O. (2013). CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 41: 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M., Farrona S., Huettel B., Koncz Z., Koncz C. (2012). CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell 24: 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare P.D., Moller S.G., Huang L.F., Chua N.H. (2003). LAF3, a novel factor required for normal phytochrome A signaling. Plant Physiol. 133: 1592–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Christner C., Kipping M., Schelbert B., Rücknagel K.P., Grabley S., Küllertz G., Fischer G. (1998). Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry 37: 5953–5960. [DOI] [PubMed] [Google Scholar]
- Heo J.B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Hernandez N. (1985). Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 4: 1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Lucito R. (1988). Elements required for transcription initiation of the human U2 snRNA gene coincide with elements required for snRNA 3′ end formation. EMBO J. 7: 3125–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.K., Shuman S. (1999). Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3: 405–411. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59. [DOI] [PubMed] [Google Scholar]
- Htun H., Lund E., Dahlberg J.E. (1984). Human U1 RNA genes contain an unusually sensitive nuclease S1 cleavage site within the conserved 3′ flanking region. Proc. Natl. Acad. Sci. USA 81: 7288–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. (2001). Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98: 8714–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. (2007). Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet. 23: 521–529. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Willingham A.T., Gingeras T.R. (2007). Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8: 413–423. [DOI] [PubMed] [Google Scholar]
- Kaul S., et al.; Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Koiwa H. (2006). Phosphorylation of RNA polymerase II C-terminal domain and plant osmotic-stress responses. In Abiotic Stress Tolerance in Plants: Toward the Improvement of Global Environment and Food, A.K. Rai and Takabe T., eds (Dordrecht, The Netherlands: Springer; ), pp. 47–57. [Google Scholar]
- Koiwa H., et al. (2002). C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl. Acad. Sci. USA 99: 10893–10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S.E., Kobor M.S., Krogan N.J., Somesh B.P., Sogaard T.M.M., Greenblatt J.F., Svejstrup J.Q. (2005). Interaction of Fcp1 phosphatase with elongating RNA polymerase II holoenzyme, enzymatic mechanism of action, and genetic interaction with Elongator. J. Biol. Chem. 280: 4299–4306. [DOI] [PubMed] [Google Scholar]
- Krishnakumar V., et al. (2015). Araport: the Arabidopsis information portal. Nucleic Acids Res. 43: D1003–D1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman T.C., Cho H., Reinberg D., Hernandez N. (1999). The general transcription factors IIA, IIB, IIF, and IIE are required for RNA polymerase II transcription from the human U1 small nuclear RNA promoter. Mol. Cell. Biol. 19: 2130–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., et al. (2014). The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet. 10: e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D., Couzigou J.M., Clemente H.S., Martinez Y., Dunand C., Bécard G., Combier J.P. (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature 520: 90–93. [DOI] [PubMed] [Google Scholar]
- Lavoie S.B., Albert A.L., Thibodeau A., Vincent M. (1999). Heat shock-induced alterations in phosphorylation of the largest subunit of RNA polymerase II as revealed by monoclonal antibodies CC-3 and MPM-2. Biochem. Cell Biol. 77: 367–374. [PubMed] [Google Scholar]
- Lerner M.R., Steitz J.A. (1979). Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 76: 5495–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M.R., Boyle J.A., Mount S.M., Wolin S.L., Steitz J.A. (1980). Are snRNPs involved in splicing? Nature 283: 220–224. [DOI] [PubMed] [Google Scholar]
- Li F., et al. (2014). Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren V., Ares M. Jr., Weiner A.M., Francke U. (1985). Human genes for U2 small nuclear RNA map to a major adenovirus 12 modification site on chromosome 17. Nature 314: 115–116. [DOI] [PubMed] [Google Scholar]
- Liu J., Gough J., Rost B. (2006). Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genet. 2: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.F., Li S.J., Chen Y., Kimberlin A.N., Cahoon E.B., Yu B. (2016). snRNA 3' end processing by a CPSF73-containing complex essential for development in Arabidopsis. PLoS Biol. 14: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine A.E., McCormick S., Estrada A., Patel K., Qin P. (2013). RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 162: 1092–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser T., Gesteland R.F. (1982). Human U1 loci: genes for human U1 RNA have dramatically similar genomic environments. Cell 29: 257–264. [DOI] [PubMed] [Google Scholar]
- Marker C., Zemann A., Terhörst T., Kiefmann M., Kastenmayer J.P., Green P., Bachellerie J.P., Brosius J., Hüttenhofer A. (2002). Experimental RNomics: identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol. 12: 2002–2013. [DOI] [PubMed] [Google Scholar]
- Matsuda O., Sakamoto H., Nakao Y., Oda K., Iba K. (2009). CTD phosphatases in the attenuation of wound-induced transcription of jasmonic acid biosynthetic genes in Arabidopsis. Plant J. 57: 96–108. [DOI] [PubMed] [Google Scholar]
- Medlin J.E., Uguen P., Taylor A., Bentley D.L., Murphy S. (2003). The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 22: 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Schell J., Koncz C. (1990). Homologous domains of the largest subunit of eucaryotic RNA polymerase II are conserved in plants. Mol. Gen. Genet. 223: 65–75. [DOI] [PubMed] [Google Scholar]
- Nelson J.D., Denisenko O., Bomsztyk K. (2006). Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1: 179–185. [DOI] [PubMed] [Google Scholar]
- Ni Z., Schwartz B.E., Werner J., Suarez J.R., Lis J.T. (2004). Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13: 55–65. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Itoh M., Okada N., Miyata T. (1981). Novel models for RNA splicing that involve a small nuclear RNA. Proc. Natl. Acad. Sci. USA 78: 4471–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M., Sugiyama M. (2005). Involvement of SRD2-mediated activation of snRNA transcription in the control of cell proliferation competence in Arabidopsis. Plant J. 43: 479–490. [DOI] [PubMed] [Google Scholar]
- Page R.D.M. (1996). TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Patturajan M., Schulte R.J., Sefton B.M., Berezney R., Vincent M., Bensaude O., Warren S.L., Corden J.L. (1998). Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273: 4689–4694. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. (2002). MicroRNAs in plants. Genes Dev. 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M.T., Voinnet O., Baulcombe D.C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski C.L., Henry R.W., Lobo S.M., Hernandez N. (1993). Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 7: 1535–1548. [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Shim E.Y., Walker A.K., Shi Y., Blackwell T.K. (2002). CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16: 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer E.J., Steitz J.A. (1993). The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996. [DOI] [PubMed] [Google Scholar]
- Sukegawa Y., Yamashita A., Yamamoto M. (2011). The fission yeast stress-responsive MAPK pathway promotes meiosis via the phosphorylation of Pol II CTD in response to environmental and feedback cues. PLoS Genet. 7: e1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A., et al. (2008). The Arabidopsis thaliana carboxyl-terminal domain phosphatase-like 2 regulates plant growth, stress and auxin responses. Plant Mol. Biol. 67: 683–697. [DOI] [PubMed] [Google Scholar]
- Uesaka M., Nishimura O., Go Y., Nakashima K., Agata K., Imamura T. (2014). Bidirectional promoters are the major source of gene activation-associated non-coding RNAs in mammals. BMC Genomics 15: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgardsdottir R., Chiodi I., Giordano M., Cobianchi F., Riva S., Biamonti G. (2005). Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell 16: 2597–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgardsdottir R., Chiodi I., Giordano M., Rossi A., Bazzini S., Ghigna C., Riva S., Biamonti G. (2008). Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 36: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arsdell S.W., Weiner A.M. (1984). Human genes for U2 small nuclear RNA are tandemly repeated. Mol. Cell. Biol. 4: 492–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetianer A., Dubois M.F., Nguyen V.T., Bellier S., Seo S.J., Bensaude O. (1995). Phosphorylation state of the RNA polymerase II C-terminal domain (CTD) in heat-shocked cells. Possible involvement of the stress-activated mitogen-activated protein (MAP) kinases. Eur. J. Biochem. 233: 83–92. [DOI] [PubMed] [Google Scholar]
- Vera J.M., Dowell R.D. (2016). Survey of cryptic unstable transcripts in yeast. BMC Genomics 17: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A., Passarelli M.C., Yario T.A., Tycowski K.T., Steitz J.A. (2015). Widespread inducible transcription downstream of human genes. Mol. Cell 59: 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waibel F., Filipowicz W. (1990). RNA-polymerase specificity of transcription of Arabidopsis U snRNA genes determined by promoter element spacing. Nature 346: 199–202. [DOI] [PubMed] [Google Scholar]
- Walker A.K., Boag P.R., Blackwell T.K. (2007). Transcription reactivation steps stimulated by oocyte maturation in C. elegans. Dev. Biol. 304: 382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.B., Brendel V. (2004). The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 5: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang X., Deng W., Fan X., Liu T.T., He G., Chen R., Terzaghi W., Zhu D., Deng X.W. (2014). Genomic features and regulatory roles of intermediate-sized non-coding RNAs in Arabidopsis. Mol. Plant 7: 514–527. [DOI] [PubMed] [Google Scholar]
- Wani S., Yuda M., Fujiwara Y., Yamamoto M., Harada F., Ohkuma Y., Hirose Y. (2014). Vertebrate Ssu72 regulates and coordinates 3'-end formation of RNAs transcribed by RNA polymerase II. PLoS One 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Shatkin A.J. (1999). Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 13: 1774–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G., Zabielski J., Hammarström K., Monstein H.J., Bark C., Pettersson U. (1984). Clustered genes for human U2 RNA. Proc. Natl. Acad. Sci. USA 81: 3811–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41: 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J.C., Dufour M.E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Libri D., Jacquier A. (2005). Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. [DOI] [PubMed] [Google Scholar]
- Xiang C., Han P., Lutziger I., Wang K., Oliver D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40: 711–717. [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Lee H., Ishitani M., Tanaka Y., Stevenson B., Koiwa H., Bressan R.A., Hasegawa P.M., Zhu J.K. (2002). Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10899–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutani I., Ozawa S., Nishida T., Sugiyama M., Komamine A. (1994). Isolation of temperature-sensitive mutants of Arabidopsis thaliana that are defective in the redifferentiation of shoots. Plant Physiol. 105: 815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Thieme C.J., Kollwig G., Apelt F., Yang L., Winter N., Andresen N., Walther D., Kragler F. (2016). tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Guo X., Ge C., Ma Z., Jiang M., Li T., Koiwa H., Yang S.W., Zhang X. (2017). KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: 4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]