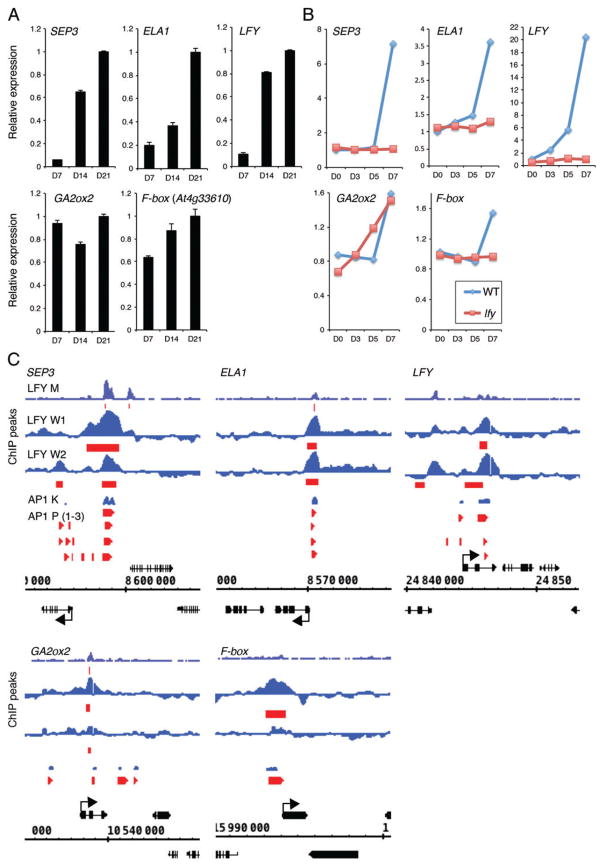
Fig. 4.
High-confidence direct LFY and AP1 upregulated targets during the switch to flower formation. (A) Expression of high-confidence direct LFY and AP1 target genes at different times of development and in different genetic backgrounds based on a public transcriptome dataset (Schmid et al. 2005). For criteria used to identify these genes, see Table S6. (B) Induction of high-confidence direct LFY and AP1 target genes by long-day photoperiod in wild-type or lfy mutant inflorescences based on a public transcriptome dataset (Schmid et al. 2003). (C) LFY and AP1 binding peaks to the regulatory regions of high-confidence direct LFY and AP1 target genes on the basis of chromatin immunoprecipitation. Screenshots were taken from the Integrated Genome Browser (IGB; http://bioviz.org/igb/) using available data for LFY [LFY M (Moyroud et al. 2011); LFY W1, W2 (Winter et al. 2011)]. Data for AP1 [AP1 K (Kaufmann et al. 2010)] were obtained through IGB (Nicol et al. 2009) using the PRI-CAT data source (http://www.ab.wur.nl/pricat/tutorial.html). AP1 K: replicate 1 data are displayed. AP1 P (1–3) (Pajoro et al. 2014): only significantly bound regions are displayed.