Figure 4. Type II-C Anti-CRISPRs Specifically Block Genome Editing by NmeCas9 in Human Cells.
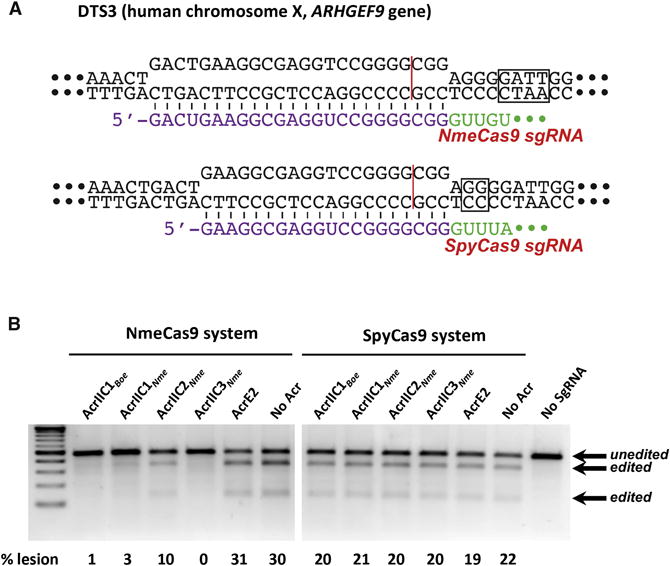
(A) Schematic representation of R-loop structures at a dual target site (DTS3) in the human genome that can be cleaved and edited by either SpyCas9 (top) or NmeCas9 (bottom). Guide sequences (purple), PAMs (boxed), and Cas9 cleavage sites (red line) are indicated.
(B) T7E1 assays of NmeCas9 or SpyCas9 editing efficiencies at DTS3 upon transient transfection of human HEK293T cells. Constructs encoding anti-CRISPR proteins were co-transfected as indicated at the top of each lane. Mobilities of T7E1-digested (edited) and -undigested (unedited) bands are indicated to the right, and editing efficiencies (“% lesion”) are given at the bottom of each lane. These images are representative of at least seven replicates.
