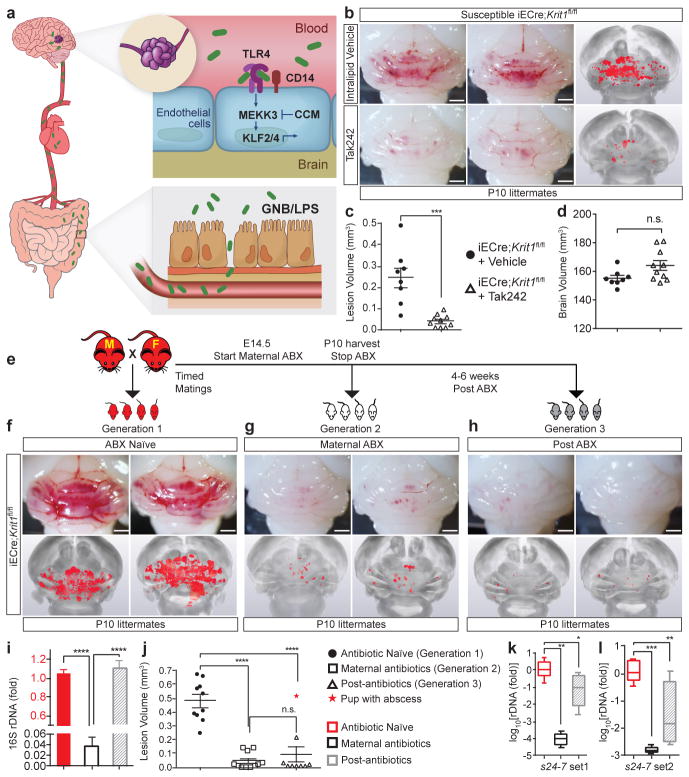
Figure 6. Preventing CCM formation by TLR4 antagonism and microbiome manipulation.
a, Model of pathogenesis in which gram negative bacteria (GNB) in the gut are the source of LPS that enters circulating blood, activating luminal, brain endothelial TLR4 receptors. LPS-TLR4 stimulation drives MEKK3-KLF2/4 signaling to induce CCMs. b, Visual and microCT images of hindbrains from vehicle or Tak242 injected animals. c–d, Lesion and brain volume quantitation. e, Intergenerational experiment in which susceptible iECre;Krit1fl/fl mating pairs were used to test the acute and chronic effects of antibiotic treatment on CCM formation. f–h, Visual and corresponding microCT images of hindbrains from offspring of three generations from one mating pair, representative of three pairs. i, Relative quantitation of neonatal gut bacterial load measured by qPCR of the bacterial 16S rRNA gene. N=4 per group. j, Lesion volume quantitation. k–l, Relative quantitation of Bacteroidetes s24-7 (s24-7) load in the neonatal gut measured by qPCR of the s24-7 rRNA gene (two distinct primer sets). N≥8 per group. All scale bars, 1 mm. Error bars shown as s.e.m. or boxplot and significance determined by one-way ANOVA with Holm-Sidak correction for multiple comparisons. ****indicates p<0.0001; ***indicates p<0.001; **indicates p<0.01; *indicates p<0.05; n.s. indicates p>0.05.