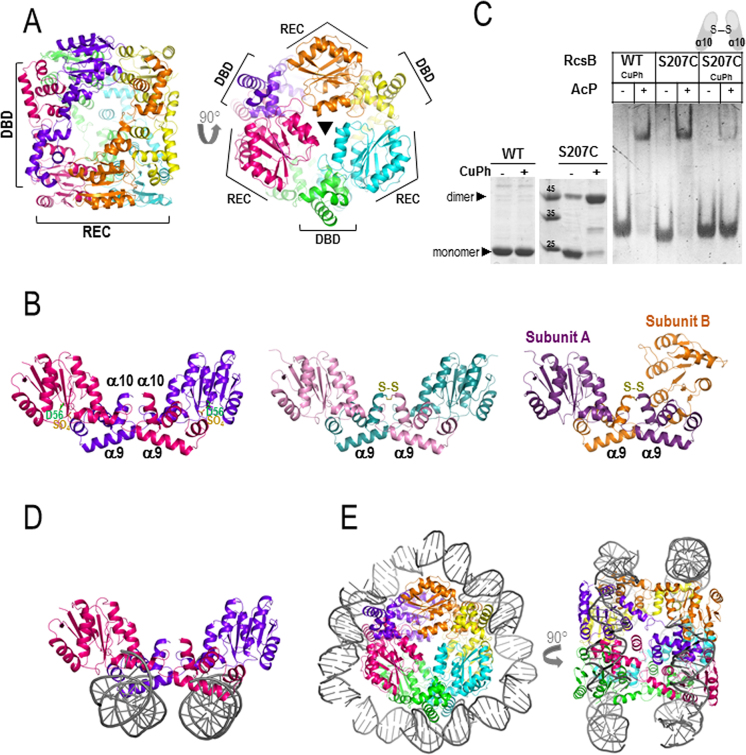
Figure 3.
Structure of RcsB in the absence of BeF3−. (A) RcsB hexamer structure, in the absence of BeF3−, composed by three dimers related by a 3-fold axis resembling a cylinder. The bases of the cylinder are generated by the REC domains and DBDs are at the central part of the cylinder. (B) Dimer crossed conformation of RcsB (RcsBcrossed; in dark pink and purple) with a sulfate ion (SO42− labeled as SO4) bound at the active site (on the left) shows dimerization mainly through REC and DBD from different subunits. Dimer structures of the S207C-RcsB mutant in the crossed conformation at the middle (S207C–RcsBcrossed; in pink and blue) and in an alternative asymmetric crossed dimer conformation on the right (S207C–RcsBAC; in violet and orange) (C) Formation of RcsBcrossed dimers with mutant S207C in the absence and presence of 0.1 mM of CuPh is observed in SDS–PAGE (left panel). EMSA experiments with WT and mutant S207C in absence and presence of 50 mM AcP and 0.1 mM CuPh (right panel) (D) In this conformation, RcsB can bind two separate dsDNA fragments (in gray) (E) Model of the RcsB hexamer wrapped by two separate dsDNA fragments.