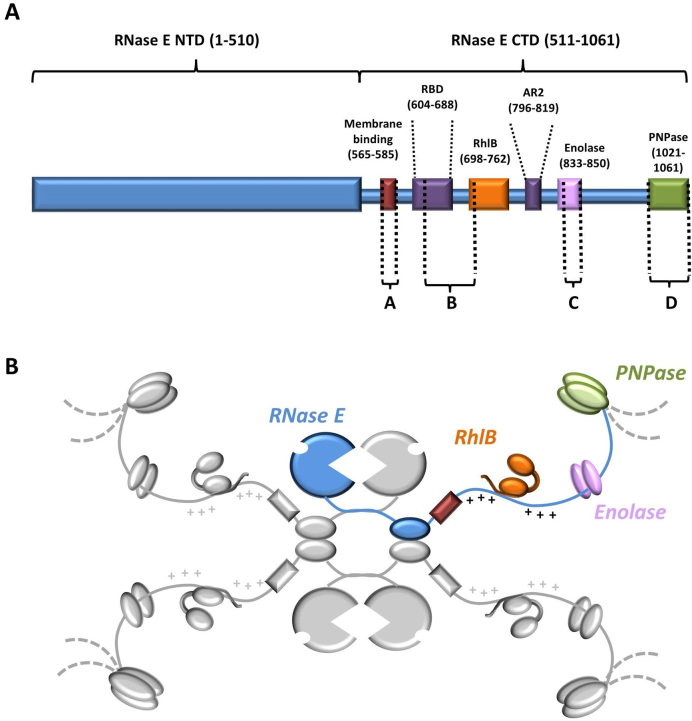
Figure 1.
Schematic representation of the Escherichia coli RNA degradosome. (A) Linear representation of RNase E with relative positions of the RNA-, protein- and membrane binding domains. Regions indicated with letters (Site A–D) have structural propensity as predicted by bioinformatics and biophysical analyses (14–16,56). (B) Cartoon of the degradosome with the tetrameric RNase E as the scaffold (one protomer in blue and the others in gray). The canonical components of the degradosome: RhlB (orange), enolase (pink) and PNPase (green) are also shown in their relative positions on the C-terminal domain of RNase E. The membrane binding domain is shown in red. The arginine-rich RNA binding domains RBD and AR2 flanking the RhlB-binding site are marked with + due to their high density of positively charged residues.