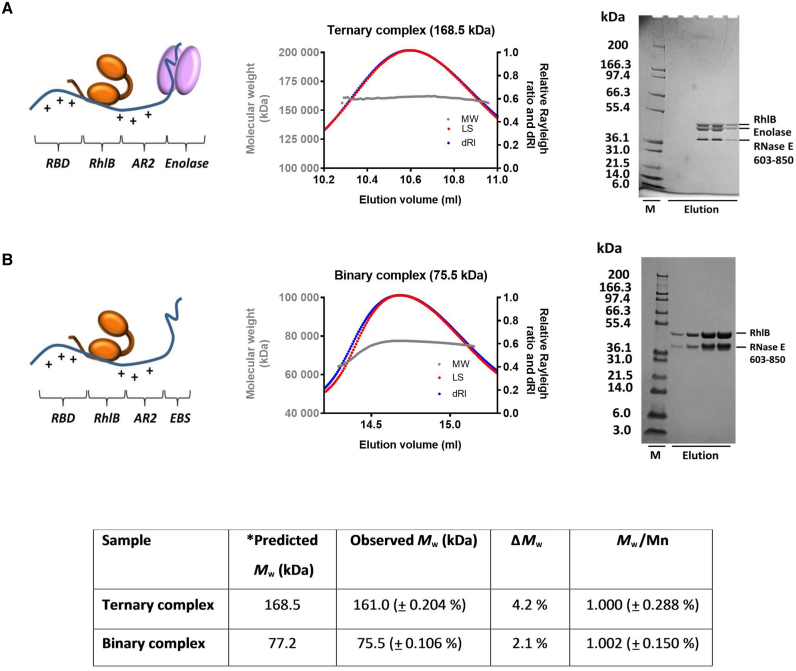
Figure 2.
Subunit composition of subassemblies of the RNA degradosome. The left panels show cartoon schematics of the subassemblies (ternary and binary complexes); the central panels show analysis by SEC-MALS in which LS (red) and differential refractive index (dRI, blue) are plotted in addition to Mw (gray); the right panels present SDS-PAGE images for the peak fractions. (A) The portion of RNase E encompassing residues 603–850 (blue) (with C-terminal Hexa-histidine tag; hereafter RNase E 603–850) interacts with RhlB (orange) and enolase (pink) to form a stable ternary complex composed RNase E 603–850: enolase: RhlB in a 1:2:1 molar ratio as indicated by the predicted and observed Mw. (B) RNase E 603–850 and RhlB form a stable binary assembly of 1:1 RNase E 603–850: RhlB. Predicted and observed Mw of the ternary and binary complexes, with their polydispersity values are shown in the table (inset). Note: RNase E 603–850 migrates slightly slower than expected, possibly due to highly charged regions or high proline content within the peptide (14,56).