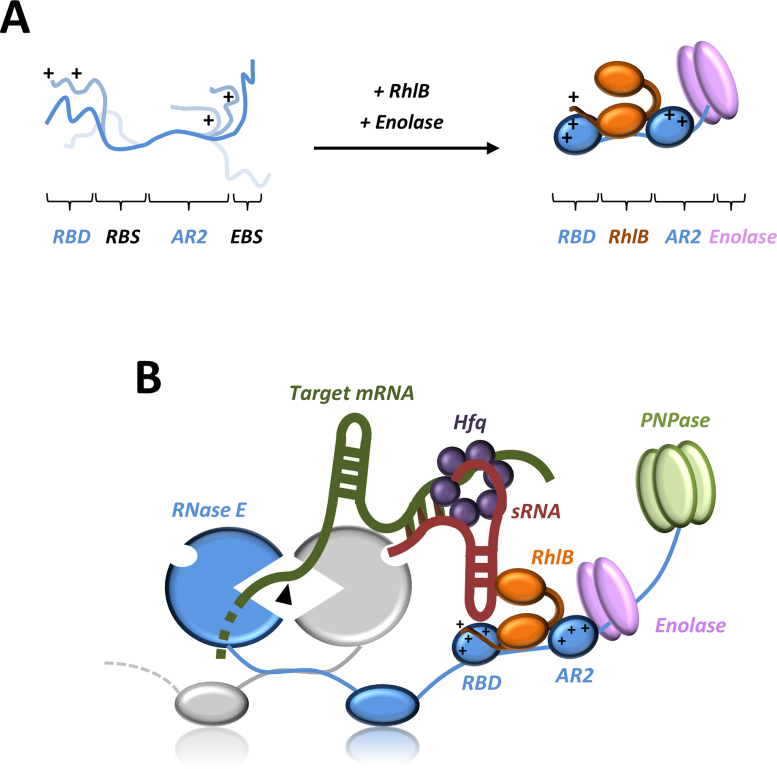
Figure 7.
Models for recognition core assembly and sRNA-mediated degradation of mRNA in the RNA degradosome. (A) RNase E 603–850 alone is highly flexible in solution, but with a preference for forming conformations which are more extended. Upon addition of RhlB and enolase, the RBD and AR2 form more limited conformations which helps pre-organize these regions for RNA binding protein and/or RNA–protein complexes. (B) A possible model for sRNA-induced degradation of target mRNA by the RNA degradosome. A sRNA–Hfq complex interacts within the recognition core region in the CTD of RNase E, via the sRNA, without displacing RhlB and enolase. Although one possible mode of interaction between the sRNA/Hfq/mRNA complex and the recognition core region in the degradosome is depicted, and we anticipate that there are various stages of interaction and remodeling in the process of sRNA-mediated degradation of target mRNA. In this case, the 5′ monophosphate end of the sRNA activates the RNase E catalytic domain by binding in the 5′ sensor domain, rendering the target mRNA susceptible to cleavage as proposed by Bandyra et al., 2012 (11).