Abstract
Yaf9 is an integral part of the NuA4 acetyltransferase and the SWR1 chromatin remodeling complexes. Here, we show that Yaf9 associates with acetylated histone H3 with high preference for H3K27ac. The crystal structure of the Yaf9 YEATS domain bound to the H3K27ac peptide reveals that the sequence C-terminal to K27ac stabilizes the complex. The side chain of K27ac inserts between two aromatic residues, mutation of which abrogates the interaction in vitro and leads in vivo to phenotypes similar to YAF9 deletion, including loss of SWR1-dependent incorporation of variant histone H2A.Z. Our findings reveal the molecular basis for the recognition of H3K27ac by a YEATS reader and underscore the importance of this interaction in mediating Yaf9 function within the NuA4 and SWR1 complexes.
INTRODUCTION
Yaf9 is a stable subunit of two major catalytic complexes acting on chromatin in Saccharomyces cerevisiae: (i) the acetyltransferase complex NuA4 that acetylates histones H4, H2A and H2A.Z and regulates transcriptional and DNA repair programs; and (ii) the ATP-dependent chromatin remodeling complex SWR1 responsible for incorporation of histone variant H2A.Z (Htz1 in yeast) into chromatin at gene promoters and required for genome stability (Figure 1A) (1–8). Yaf9 has been shown to play important roles in the functions of both complexes and is essential in the exchange of H2A-H2B for H2A.Z-H2B dimers by SWR1-C. Genetic studies with YAF9 deletion strains have implicated Yaf9 in transcriptional regulation, histone acetylation and DNA repair. Cells lacking Yaf9 are characterized by reduced telomere-proximal gene expression and sensitivity to DNA-damaging agents and stress-inducing conditions (6). In addition, Yaf9 is involved in chromosome segregation, cellular resistance to microtubule depolymerizing agents and response to spindle stress (2). The N-terminus of Yaf9 contains the YEATS (Yaf9, ENL, AF9, Taf14 and Sas5) domain (Figure 1B) that has recently been shown to bind acylated histone peptides (9–11); however the precise selectivity of Yaf9 remains unclear.
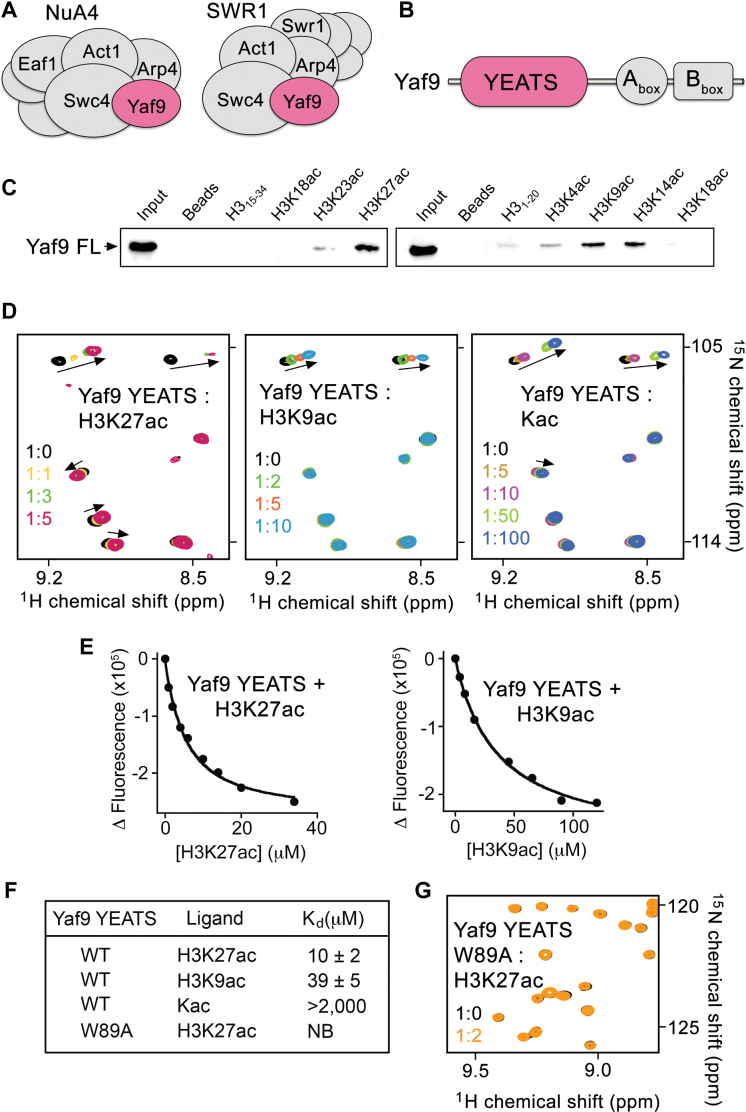
Figure 1.
The YEATS domain of Yaf9 is a reader of H3K27ac. (A) Yeast NuA4 and SWR1 complexes composition. (B) Schematic representation of the Yaf9 protein with the YEATS domain highlighted in pink. (C) Western blot analysis of pull-downs using GST-Yaf9FL and indicated biotinylated histone H3 peptides. (D) Superimposed 1H,15N heteronuclear single quantum coherence (HSQC) spectra of uniformly 15N-labeled Yaf9-YEATS with the indicated ligands added in a stepwise manner. The spectra are colored as indicated. (E) Representative binding curves used to determine Kd by intrinsic tryptophan fluorescence. (F) Binding affinities of the YEATS domain as measured by fluorescence or nuclear magnetic resonance (NMR) (for Kac). (G) Superimposed 1H,15N HSQC spectra of the 15N-labeled W89A mutant of Yaf9-YEATS in the presence and absence of H3K27ac peptide (aa 21–31 of H3).
The YEATS domain is an evolutionarily conserved module that is found in three yeast proteins (Sas5, Taf14 and Yaf9) and four human proteins (ENL, AF9, GAS41 (YEATS4) and YEATS2) (12). It is the newest member of the family of acetyllysine readers, which also includes bromodomain and the double PHD finger (DPF) (13,14). All YEATS domains appear to target acyllysine post-translational modifications (PTMs) in histones but they display differences in selectivity for the position and nature of the acyl marks. While the YEATS domains of AF9 and Taf14 have a preference for acylated, particularly crotonylated, H3K9 (9–11,15,16), the YEATS domains of ENL and YEATS2 select for acylated H3K27 (17–19). Specific recognition of acyllysine modifications mediates diverse functions of the YEATS domain-containing proteins (YEATS proteins) and ensures their involvement in non-redundant signaling pathways. Thus, binding of AF9 to H3K9ac-enriched chromatin facilitates recruitment of the methyltransferase DOT1L and enhances H3K79 methylation, while binding to H3K18cr has a role in activation of the inflammatory response genes (10,11). Interaction of Taf14 with H3K9ac is essential for transcription of target genes and DNA-damage repair in yeast (9). The ability of ENL to bind H3K27ac and H3K9ac is linked to oncogenic gene expression in acute leukemia (17,18).
In this work, we report on the structure–function activity of the YEATS domain of Yaf9 (Yaf9-YEATS). We show that this module is a selective reader of H3K27ac, as it binds to H3K27ac ∼4-fold tighter than to H3K9ac. Structural, biochemical and mutational experiments reveal that K27ac inserts between two aromatic residues that are strictly required for the interaction. Disruption of this binding in vivo impairs deposition of H2A.Z, gene transcription and the DNA-damage response. Collectively, our results and the functional relationship of H2A.Z and H3K27ac at active enhancers and promoters in higher eukaryotes (20,21) highlight both mechanistic conservation and uniqueness of reading acetyllysine marks by the YEATS domains. Our findings further shed light on the significance of this interaction for biological activities of Yaf9 in yeast and its ortholog GAS41/YEATS4 in the mammalian TIP60/p400 complex (1,22) and underscore integrated functions of the NuA4 and SWR1-C complexes.
MATERIALS AND METHODS
Cloning, mutagenesis and protein purification
The wild-type (WT) YEATS domains of Yaf9 (aa 8–171) and Taf14 (aa 1–137) were cloned into pGEX-6P1 vectors. WT AF9 YEATS domain (aa 1–138) in a pET28b vector was a kind gift from Haitao Li. The YEATS domain mutants of Yaf9 (W89A, E91G, R114D), Taf14 (G83E, D104R) and AF9 (G80E) were generated using the Stratagene QuikChange Lightning Site Directed Mutagenesis Kit. The WT and mutant YEATS domains were expressed and purified using the previously described protocol (16). Briefly, all proteins were expressed in Escherichia coli Rosetta2 (DE3) pLysS or BL21 (DE3) RIL in either LB or minimal media supplemented with 15NH4Cl. Protein expression was induced with 0.3–1.0 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells were grown at 16–18°C overnight, spun down and resuspended in lysis buffer (20–50 mM Tris–HCl or Hepes pH 7.5–8.0, 150–500 mM NaCl, 5 mM dithiothreitol or 2 mM BME or 1 mM TCEP, and 0.1–1% Trition X-100). Resuspended cells were flash-frozen in liquid N2 and stored at −80°C. Cells were thawed and sonicated on ice and cellular debris was removed by centrifugation. The GST-fusion proteins were purified on glutathione Sepharose 4B beads (Thermo Fisher Sci), and the GST-tag was cleaved using PreScission protease overnight at 4°C. The His-tag fusion proteins were purified using nickel–NTA resin (Qiagen), and the His tag was cleaved by thrombin. Proteins were further purified by size exclusion chromatography using a S100 column or HiTrap SP HP and HiPrep 16/600 Superdex 75 columns (GE Healthcare). The WT GAS41-YEATS (aa 9–151) construct was generated and the proteins were purified as above.
NMR spectroscopy
Nuclear magnetic resonance (NMR) experiments were performed at 298 K on a Varian INOVA 600 MHz spectrometer equipped with a cryogenic probe. 1H,15N heteronuclear single quantum coherence (HSQC) spectra of 0.1–0.2 mM uniformly 15N-labeled WT or mutated Yaf9 (in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM TCEP, 8–10% D2O), Taf14 (in 1× phosphate buffered saline (PBS) pH 6.8, 1 mM TCEP, 8–10% D2O) and AF9 (in 25 mM Tris–HCl pH 7.5, 250 mM NaCl, 2 mM BME, 8–10% D2O) YEATS domains were collected to confirm that all the proteins are folded. 1H,15N HSQC spectra of the Yaf9 YEATS domain were recorded in the presence of increasing concentration of H3K27ac (aa 21–31 of H3) or H3K9ac (aa 5–13 of H3) (synthesized by Synpeptide) or free acetyllysine. Kd value was calculated by a nonlinear least-squares analysis in Kaleidagraph using the equation:
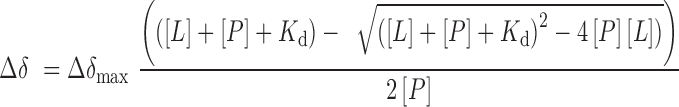 |
where, [L] is concentration of free acetyllysine, [P] is concentration of the protein, Δδ is the observed chemical shift change and Δδmax is the normalized chemical shift change at saturation. Normalized chemical shift changes were calculated using the equation 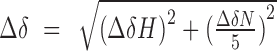 , where Δδ is the change in chemical shift in parts per million.
, where Δδ is the change in chemical shift in parts per million.
Fluorescence spectroscopy
Spectra were recorded at 25°C on a Fluoromax-3 spectrofluorometer (HORIBA). The samples containing the WT and mutated YEATS domains of Yaf9 (in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM TCEP), Taf14 (in 1× PBS pH 7.4), or AF9 (in 25 mM Tris–HCl pH 7.5, 500 mM NaCl, 2 mM BME) and progressively increasing concentrations of the histone H3K27ac (aa 21–31 of H3) or H3K9ac (aa 5–13 of H3) peptides were excited at 295 or 280 nm. Emission spectra were recorded over a range of wavelengths between 310 and 400 nm with a 0.5–0.6 nm step size and 1 s integration time. The Kd values were determined using a non-linear least-squares analysis and the equation:
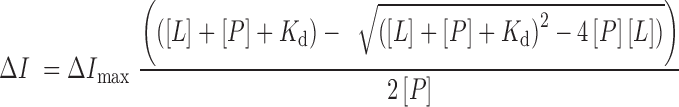 |
where, [L] is the concentration of the histone peptide, [P] is the concentration of the protein, ΔI is the observed change of signal intensity and ΔImax is the difference in signal intensity of the free and bound states of the protein. The Kd values were averaged over two or three separate experiments with error calculated as the standard deviation between the runs.
X-Ray crystallography
Purified Yaf9-YEATS (aa 8–171) at 14 mg/ml in 20 mM Tris–HCl pH 7.5, 150 mM NaCl and 1 mM TCEP was incubated on ice with a 5 molar equivalence of H3K27ac peptide (21–31 aa) for 1–2 h. Crystals of the protein:peptide complex were grown using sitting-drop vapor diffusion against an equal volume of well solution containing 2.0 M ammonium sulfate, 0.1 M Bis-Tris pH 5.5 at 4°C. Crystals were cryoprotected with the addition of 30% glycerol to the well solution and the X-ray diffraction data were collected on the UC Denver X-ray Crystallography core facility Rigaku Micromax 007 high-frequency microfocus X-ray generator equipped with a Pilatus 200K 2D area detector. Indexing and scaling were completed using HKL3000. The phase solution was solved with Phenix.phaser using molecular replacement and the apo- Yaf9 YEATS (PDB ID: 3FK3) structure as the model. Model building was carried out with Coot and refinement was performed with Phenix.refine (23,24). The final structure was validated with MolProbity (25). Crystallographic statistics for the Yaf9 YEATS–H3K27ac structure are shown in Supplementary Table S1.
Yeast strains and growth assay
Standard growth condition, media and manipulation techniques were used for the construction of strains used in this study. Genomic DNA was used to amplify YAF9 ORF which was cloned in zero blunt TOPO vector (Invitrogen). The point mutations were introduced in this construct by polymerase chain reaction (PCR). The insert, KanR and NatR cassettes were then amplified and transformed in yeast cells as described (26). The alleles were inserted in the BY4741 and QY708 backgrounds (26).
The yeast spot assays were done in BY4741 background with yeast culture grown overnight in YPD at 30°C, inoculated in the morning in YPD at OD600 0.3 and grown for 3–4 h. The 10-fold serial dilutions of the yeast cultures were made for spotting on YPD plates supplemented with methylmethane sulfonate (MMS)-0.025% or formamide-3% and the growth sensitivities were compared to YPD control plate.
Tandem affinity purification (TAP) of NuA4 complex
TAP purification was performed as described in (26) with the following modifications. Yeast cells were grown in 250 ml of YPD overnight until OD600 1.5. After TEV treatment the elution was incubated with Protein A bound sepharose beads to remove any leaked IgG, for 20 min on a wheel at 4°C. Western analysis and antibodies used have been described in (1).
GST purification and pull-down experiments
Standard procedures were used to purify recombinant GST fused YEATS domain of Yaf9 described in (27). Following IPTG induction the bacterial culture was grown overnight at 16°C, lysed with lysozyme treatment followed by sonication. The cell lysate was incubated with glutathione sepharose beads for 4 h at 4°C. For the peptide pull-down experiment, the recombinant proteins were eluted with glutathione whereas, for chromatin pull-down experiment, the recombinant proteins were not eluted and the beads were stored at 4°C.
The biotinylated peptides were obtained from AnaSpec. One microgram of peptide was incubated with 2 μg of recombinant proteins overnight on a wheel at 4°C in binding buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% NP-40, 1 mM PMSF). Next morning the protein/peptide solution was mixed with streptavidin dynabeads for 2 h at 4°C, washed three times followed by western blotting with anti-Yaf9 (Abcam) and anti-GST (Sigma).
Chromatin pull-down assay was performed with 5 μg of purified mammalian H1-depleted oligonucleosomes incubated with 2.5 μg of immobilized GST fusion recombinant proteins overnight at 4°C in binding buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% NP-40, 1 mM PMSF). After washes in the same buffer, western blotting was performed with anti-H3K27ac (Abcam) and anti-H4 (Abcam).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) experiments were performed as described previously (26). Cells were grown in YPD to an O.D. between 0.5 and 1. The cells were cross-linked for 20 min using 1% formaldehyde. Chromatin was sonicated (Diagenode Bioruptor) to get 200–500 bp fragments. A total of 100 μg of chromatin was used for immunoprecipitation with anti-Htz1 C-term (Millipore) and anti-H4 (Abcam). DNA was quantified on LC480 LightCycler (Roche) using primer pairs to evaluate occupancy at the PHO5 promoter (corresponding to the UAS2 element) (% of immunoprecipitation/input). The data were corrected for nucleosome occupancy by using total H4 signal. The values are based on two biological replicates.
RESULTS AND DISCUSSION
The YEATS domain of Yaf9 is a preferential reader of H3K27ac
To establish selectivity of Yaf9 for acetyllysine modifications in histone H3, we produced the GST-Yaf9 construct and tested it by in-solution peptide pull-down assays (Figure 1C). We found that GST-Yaf9 preferably binds to the histone H3K27ac peptide. It also associated with H3K9ac and H3K14ac, though to a lesser extent, but did not recognize other acetylated PTMs on H3 (Figure 1B). As the sequence of H3 surrounding K9 and K27 is similar, we particularly focused on determining the mechanistic basis underlying the difference in recognition of these two critical epigenetic marks.
The preference of Yaf9-YEATS for H3K27ac was substantiated by NMR and fluorescence spectroscopy data. Titration of the H3K27ac peptide (residues 21–31 of H3) to uniformly 15N-labeled Yaf9-YEATS led to substantial chemical shift perturbations (CSPs) in 1H,15N HSQC spectrum of the protein, implying direct binding. A number of resonances corresponding to the free state of Yaf9-YEATS moved and broadened revealing an intermediate exchange regime on the NMR timescale that is indicative of a tight interaction (Figure 1D, left panel). In agreement, a 10 μM binding affinity of Yaf9-YEATS for the H3K27ac peptide was measured by intrinsic tryptophan fluorescence (Figure 1E and F). However gradual addition of H3K9ac (residues 5–13 of H3) to the Yaf9-YEATS NMR sample caused CSPs in the intermediate-to-fast exchange regime. This pattern of CSPs suggested a significant decrease in binding (Figure 1D, middle panel), and in support, the determined equilibrium dissociation constant (Kd) of 39 μM confirmed that binding of Yaf9-YEATS to H3K9ac is ∼4-fold weaker compared to its binding to H3K27ac (Figure 1E and F). Together, these results demonstrate that the YEATS domain of Yaf9 is a selective reader of the H3K27ac mark, which is deposited by the acetyltransferase Gcn5/SAGA in yeast (28).
Structural mechanism for the recognition of H3K27ac
To gain mechanistic insight into the interaction, we obtained a crystal structure of the Yaf9 YEATS domain in complex with the H3K27ac peptide (Figure 2 and Supplementary Table S1). The crystallographic asymmetric unit was comprised of four copies of the complex (Supplementary Figure S1). The Yaf9 YEATS domain folds into a canonical immunoglobin β-sandwich structure, which consists of eight β-strands connected by variable loops and capped by a short α-helix at one of the open ends of the β-sandwich. The loops at the opposite end of the β-sandwich form a binding site for the acetyllysine. The H3K27ac peptide lays perpendicular to the β-strands with the side chain of K27ac, being in an extended conformation, crossing the β-sandwich at a right angle (Figure 2A and B). The acetyl group of K27ac is inserted between two aromatic residues, Y70 and W89, and is constrained through a hydrogen bond formed between its carbonyl oxygen and the backbone amide of W89 and via water-mediated hydrogen bonds with the backbone carbonyl group of G90 and the hydroxyl group of Y70 (Figure 2C and D). The ε nitrogen atom of K27ac donates a hydrogen bond to the hydroxyl group of T69. In addition, a set of polar interactions involving residues 28–31 of the histone peptide stabilizes the complex. Characteristic main chain β-sheet hydrogen bonding contacts are made between S28 of the peptide and E91 of the protein, and the backbone amide of A31 provides further stabilization of the peptide via a water-mediated hydrogen bond with the carboxyl group of E91. The contacts with the peptide residues besides K27ac are essential as binding of free acetyllysine amino acid was substantially, over 200-fold, weaker (Figure 1D, right panel and Figure 1F).
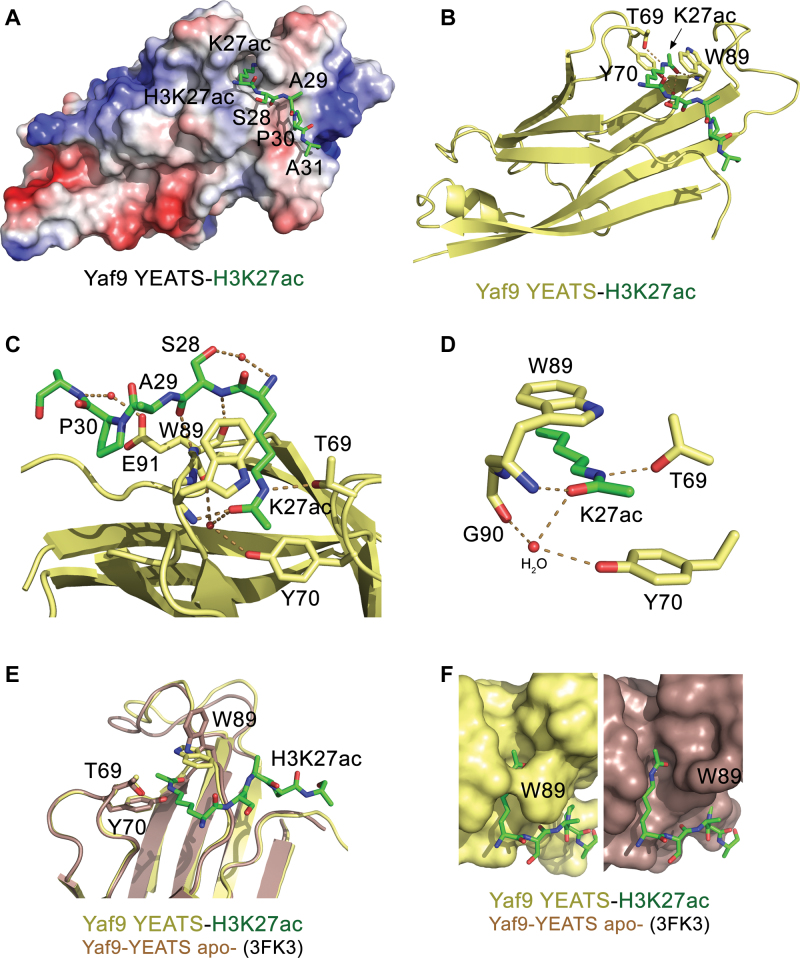
Figure 2.
Structural mechanism for the recognition of H3K27ac. (A) The electrostatic surface potential of Yaf9-YEATS is colored blue and red for positive and negative charges, respectively. Bound H3K9ac peptide is shown as green sticks. (B) The ribbon diagram of the Yaf9 YEATS–H3K27ac complex. Dashed lines represent hydrogen bonds and red spheres represent water molecules. The YEATS domain is colored yellow and H3K27ac peptide is colored green. Residues of the YEATS domain involved in the binding of K27ac are labeled. (C and D) Close up view of the H3K27ac-binding pocket of Yaf9-YEATS. (E) An overlay of the structures of Yaf9-YEATS in the H3K27ac bound state (yellow/green) and the apo-state (PDB ID: 3FK3, brown). (F) A zoom-in view of the K27ac-binding pocket in the complex and in the apo-state. Note, H3K27ac peptide is manually positioned in the binding groove of the apo-state.
Overall the structure of the H3K27ac-bound YEATS domain of Yaf9 superimposes well with the structure of Yaf9-YEATS in the apo-state (PDB ID: 3FK3) (5) (root mean square deviation (rmsd) = 0.3 Å). The indolyl moiety of W89 in two apo-state YEATS molecules (out of three in one asymmetric unit), is positioned away from the K27ac-binding groove (Figure 2E and F), but it adopts the same position as in the complex in the third molecule, flipping over the groove and creating a tunnel where acetyllysine is locked in place. Substitution of W89 with an alanine abolished binding of Yaf9-YEATS to H3K27ac, underscoring the imperative role of the aromatic cage in the recognition of this PTM (Figure 1G).
Binding to H3K27ac is important for Yaf9 function within NuA4 and SWR1-C complexes
To define the functional significance of the Yaf9 YEATS domain, we generated Y70A and W89A mutants of Yaf9 and examined them in cell assays. We found that neither mutant affects Yaf9 association with its protein complexes in vivo, unlike deletion of C-terminal box B (ΔC, Yaf9 (residues 1–186)), corroborating results reported previously (5,6) (Figure 3A). Peptide and nucleosome pull-down assays with Yaf9-YEATS confirmed that the W89A mutation disrupts recognition of H3K27ac in vitro (Figure 3B and C). Strikingly, the substitution mutants showed growth phenotypes on specific media/conditions similar to the full length YAF9 gene deletion or C-terminal truncation, revealing a critical role of Yaf9-YEATS in genome expression and stability (Figure 3D).
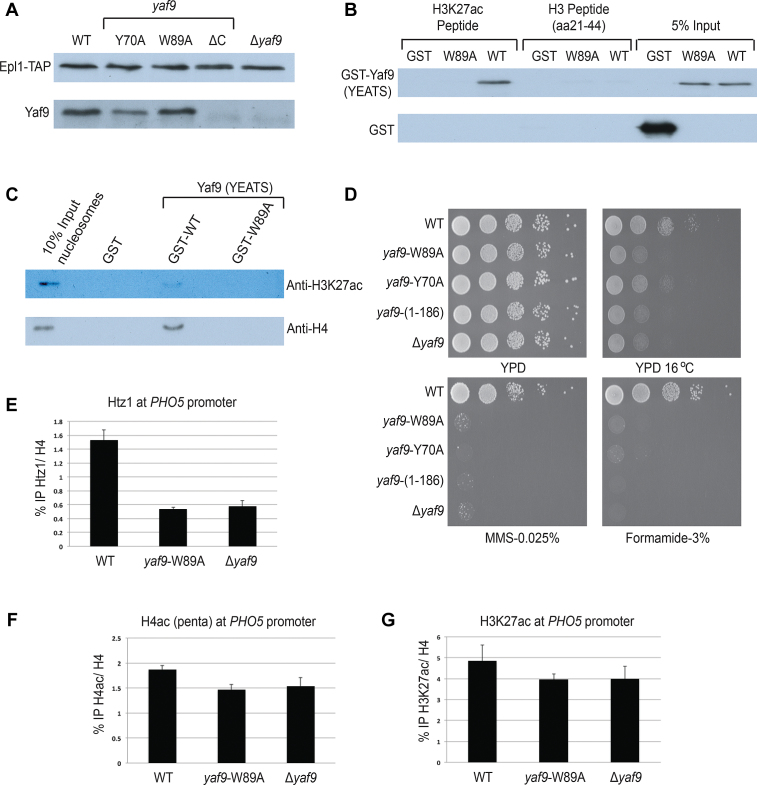
Figure 3.
Mutations in Yaf9-YEATS disrupt binding to H3K27ac in vitro and loading of H2A.Z in vivo. (A) TAP purification of NuA4 complex using Epl1-TAP tagged background. Yaf9 WT and point mutants (Y70A and W89A) associate with the NuA4 complex, but the C-terminal truncated form (ΔC; 1–186 aa) of Yaf9 does not. Purified NuA4 complex was ran on 10% Sodium dodecyl sulphate-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membrane and detected with indicated antibodies. (B) Peptide pull-down assay using indicated H3 peptides and GST-tagged Yaf9-YEATS. Recombinant WT Yaf9 binds to H3K27ac peptides, whereas the W89A mutant does not. None of the proteins show binding to unmodified H3 peptide. Empty GST is used as negative control. (C) Chromatin pull-down assay using human nucleosomes and GST-tagged Yaf9-YEATS. WT Yaf9 binds to nucleosomes harboring the H3K27ac mark, while the W89A mutant does not. Empty GST is used as negative control. (D) Phenotypic analysis of yeast strains with integrated alleles YAF9 (WT), yaf9-Y70A, yaf9-W89A, yaf9(1–186) and Δyaf9. The strains expressing the YEATS domain point mutants showed similar growth defects as truncated yaf9(1–186) and Δyaf9 at 16°C, and in the presence of formamide or DNA-damaging agent MMS. (E) The point mutation W89A introduced in endogenous Yaf9 creates a strong defect in incorporation of histone variant H2A.Z (Htz1) at the PHO5 gene promoter in vivo. ChIP-qPCR was performed using anti-Htz1 and anti-H4 antibodies and the indicated yeast strains. The precipitated DNA was quantified with primers spanning the PHO5 UAS2 region. Data are presented as ratio of IP for Htz1 normalized on total H4. The error bar represents range between two biological replicates. (F and G) Levels of H4 hyperacetylation and H3K27ac were measured by ChIP-qPCR in the same conditions as in (E).
Yaf9 is known to be required for H2A.Z incorporation into chromatin both in vitro and in vivo (1,5,6,29,30). We performed ChIP-qPCR analysis at the PHO5 gene, which is tightly regulated by H2A.Z (Htz1), NuA4 and Gcn5/SAGA HAT complexes (1,31–33). As shown in Figure 3E, H2A.Z occupancy is clearly lost at the PHO5 promoter in cells expressing the Yaf9-W89A mutant, similar to what is observed with the full Yaf9 deletion. Importantly, H3K27ac was also enriched at the PHO5 promoter but was not significantly affected by Yaf9 mutation, while NuA4-dependent H4 acetylation may be slightly decreased (Figure 3F and G). Our results indicate that recognition of H3K27ac-containing nucleosomes by the YEATS domain of Yaf9, associated with the SWR1 complex, leads to the replacement of H2A-H2B dimers with H2A.Z-H2B at the PHO5 promoter. Interestingly, H3K27ac and H2A.Z are chromatin hallmarks known to co-localize at activating gene regulatory elements in higher eukaryotes (enhancers and promoters) (20,21,34).
Conservation and uniqueness of the H3K27ac-recognition mechanism
Comparison of the structures of the Yaf9 YEATS domain in complex with H3K27ac (hereafter referred to as Yaf9–H3K27ac) and the corresponding complexes Taf14–H3K9ac (PDB ID: 5D7E) (9,16) and YEATS2-H3K27cr (histone H3 crotonylated on lysine 27) (PDB ID: 5IQL) (19) reveals a unique acyllysine-recognition mechanism that is conserved in Yaf9 and YEATS2 but differs substantially from the binding mechanism used by Taf14 (Figure 4). While Taf14 recognizes primarily the histone H3 sequence N-terminal to the acyllysine mark (K9acyl), including Q5-R8, both Yaf9 and YEATS2 are in contact primarily with the residues C-terminal to the acyllysine mark (K27acyl) (9,15,16,19). An overlay of the structures of Yaf9–H3K27ac and YEATS2-H3K27cr (rmsd of 0.6 Å) showed that the position of K27acyl is strictly conserved in both complexes, and that the S28-A31 sequence of H3K27ac peptide occupies the respective shallow hydrophobic-binding pockets in a similar way (Figure 4B). On the other hand, little-to-no contacts are seen with the peptide residues C-terminal to K27acyl in both complexes: the R26 residue is entirely solvent exposed in YEATS2-H3K27cr and we did not observe clear electron density for this residue in Yaf9–H3K27ac.
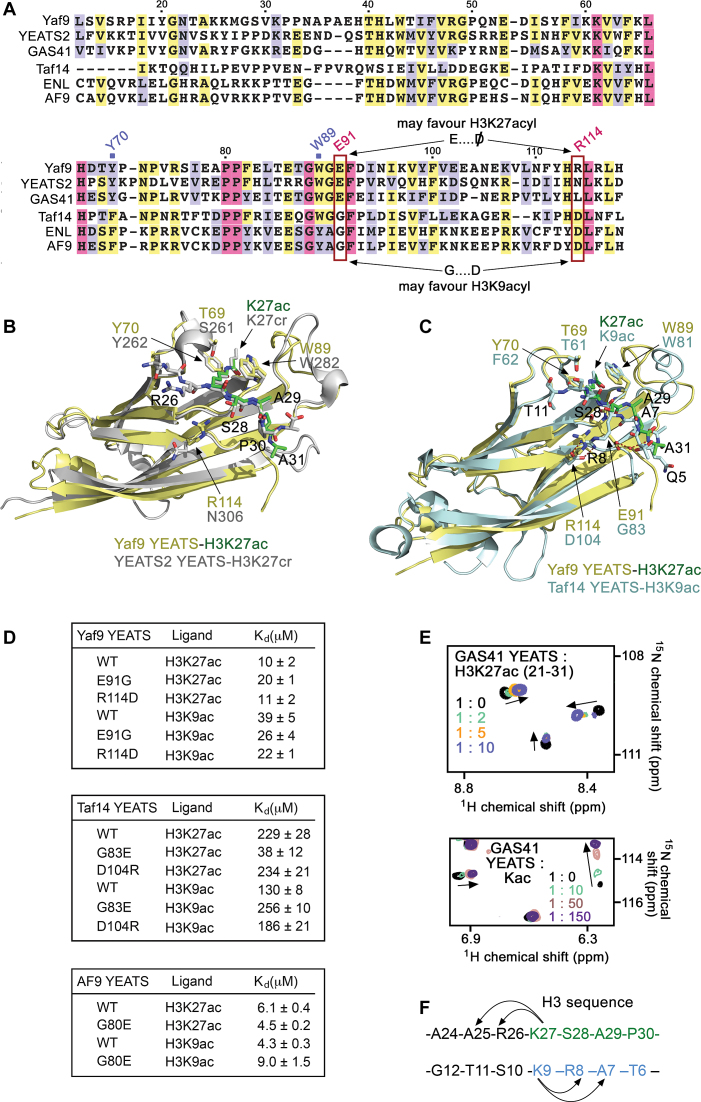
Figure 4.
Analysis of the H3K27ac-recognition mechanism. (A) Alignment of the YEATS domain sequences: absolutely, moderately and weakly conserved residues are colored pink, yellow and lavender, respectively. (B and C) Superimposed structures of Yaf9–H3K27ac (yellow/green), YEATS2-H3K27cr (PDB ID: 5IQL, gray), and Taf14–H3K9ac (PDB ID: 5D7E, cyan). (D) Binding affinities of the indicated WT and mutated YEATS domains as measured by fluorescence. (E) Superimposed 1H,15N HSQC spectra of GAS41-YEATS recorded in the presence of increasing concentration of H3K27ac peptide or Kac. Spectra are color-coded according to the protein:ligand molar ratio. (F) Sequence of H3 with the region recognized by Yaf9-YEATS and YEATS2-YEATS colored green, and the region recognized by AF9-YEATS and Taf14-YEATS colored blue. The arrows represent similar positions of the histone H3 residues around K27 and K9.
By contrast, the Taf14 YEATS domain exhibits preference for the H3K9acyl peptides, which in the Taf14–H3K9acyl complexes are bound in an orientation that is opposite to the orientation of the H3K27acyl peptides in Yaf9–H3K27ac and YEATS2-H3K27cr (Figure 4C and F). It has previously been shown that in addition to the recognition of acyllysine, which is undoubtedly the main driving force of the interaction, yeast Taf14 and the orthologous human protein AF9 display selectivity toward an RKacyl motif (9–11,16,35). The guanidino group of R8, the residue preceding acyllysine (K9acyl), forms a salt bridge with an aspartate that is conserved in Taf14 and AF9 (D104 in Taf14). Formation of the R-D salt bridge has a substantial impact on the interaction, as mutation of R8 to an alanine decreases binding of AF9 ∼200-fold (11). However, Yaf9 contains R114 and YEATS2 contains an asparagine instead of the aspartate, which not only leads to elimination of the salt bridge, but may also result in the repulsion of the RKacyl motif in the case of Yaf9 (Figure 4A, red box). Indeed, when R114 in Yaf9-YEATS was substituted with an aspartate, the binding activity to H3K9ac was increased from 39 μM (WT Yaf9) to 22 μM (Yaf9 R114D), whereas binding to H3K27ac was unaffected (Figure 4D). In further support of the importance of aspartate for the interaction with H3K9ac, the reciprocal mutation in Taf14-YEATS, D104R, also led to a decrease in binding to H3K9ac but not to H3K27ac (Figure 4D).
In the Yaf9–H3K27ac complex, E91 of the protein forms stabilizing hydrogen bonds with the H3K27ac peptide; however E91 might produce steric hindrance for binding of the H3K9ac peptide, as seen from an overlay of the structures of the Yaf9–H3K27ac and Taf14–H3K9ac complexes (Figure 4C). To explore the effect of E91 in Yaf9 and of the corresponding G83 in Taf14, we generated the E91G mutant of Yaf9-YEATS and the G83E mutant of Taf14-YEATS and examined their interactions by tryptophan fluorescence (Figure 4D). Remarkably, we found that the G83E mutant of of Taf14-YEATS altered its selectivity and favored H3K27ac instead of H3K9ac, which is a preferred ligand of WT Taf14-YEATS. The binding of G83E Taf14-YEATS to H3K27ac was increased ∼6-fold, whereas its binding to H3K9ac was decreased ∼2-fold. Furthermore, binding of the reciprocal E91G mutant of Yaf9-YEATS to H3K27ac was decreased ∼2-fold but its binding to H3K9ac was increased ∼1.5-fold. Even though the Yaf9-YEATS domain did not flip its selectivity as Taf14-YEATS did, the fact that the E91G Yaf9-YEATS bound equally well to H3K9ac and H3K27ac is in full agreement with the idea that the residue in the position +2 with respect to the aromatic cage tryptophan (W89 in Yaf9) has an essential role in selectivity of the YEATS domain. We confirmed this again by replacing the respective G80 in the AF9 YEATS domain with a glutamate: while WT AF9-YEATS slightly selected for H3K9ac, the G80E mutant of AF9-YEATS switched its preference to H3K27ac (Figure 4D).
GAS41/YEATS4 is often referred to as ortholog of Yaf9 in higher eukaryotes because it resides in the SRCAP and TIP60/p400 complexes, corresponding to SWR1 and the NuA4/SWR1 physical merge in yeast (36,37). Much like the sequences of Yaf9 and YEATS2, the sequence of GAS41-YEATS contains a glutamate at the +2 position to the conserved Trp and lacks the salt-bridge forming aspartate, suggesting that GAS41 might also recognize H3K27acyl modification (Figure 4A). Although the structure of GAS41-H3K27acyl is required to define the orientation of H3K27acyl peptide in the complex, NMR titration experiments indicate a tight binding of GAS41-YEATS to H3K27ac and that along with K27ac, other residues of the histone peptide are involved in the interaction with the YEATS domain (Figure 4E).
Concluding remarks
In this study, we show structural determinants underlying acetyllysine selectivity of the YEATS domains that target H3K27ac. This led us to establish a novel functional link between H3K27ac and the deposition of histone variant H2A.Z at gene regulatory elements. While H2A.Z is often linked to poised promoters, its co-localization with H3K27ac correlates with activation. Therefore, our data provide molecular insight into how these two chromatin hallmarks can be coupled during the process of gene activation.
AVAILABILITY
Atomic coordinates for the structure of the Yaf9 YEATS–H3K27ac complex have been deposited in Protein Data Bank under accession code 6AXJ.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) Grants [R01 GM106416, GM125195, GM100907 to T.G.K., GM110058 to B.D.S.]; CIHR Grant [FDN-143314 to J.C.]; Canada Research Chair in Chromatin Biology and Molecular Epigenetics (J.C.). Funding for open access charge: NIH [GM100907].
Conflict of interest statement. None declared.
REFERENCES
- 1. Auger A., Galarneau L., Altaf M., Nourani A., Doyon Y., Utley R.T., Cronier D., Allard S., Cote J.. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol. Cell. Biol. 2008; 28:2257–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Le Masson I., Yu D.Y., Jensen K., Chevalier A., Courbeyrette R., Boulard Y., Smith M.M., Mann C.. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell. Biol. 2003; 23:6086–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu P.Y., Levesque N., Kobor M.S.. NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components. Biochem. Cell Biol. 2009; 87:799–815. [DOI] [PubMed] [Google Scholar]
- 4. Millar C.B., Xu F., Zhang K., Grunstein M.. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006; 20:711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang A.Y., Schulze J.M., Skordalakes E., Gin J.W., Berger J.M., Rine J., Kobor M.S.. Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:21573–21578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang H., Richardson D.O., Roberts D.N., Utley R., Erdjument-Bromage H., Tempst P., Cote J., Cairns B.R.. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol. Cell. Biol. 2004; 24:9424–9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krogan N.J., Baetz K., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R. et al. . Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Altaf M., Auger A., Monnet-Saksouk J., Brodeur J., Piquet S., Cramet M., Bouchard N., Lacoste N., Utley R.T., Gaudreau L. et al. . NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 2010; 285:15966–15977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shanle E.K., Andrews F.H., Meriesh H., McDaniel S.L., Dronamraju R., DiFiore J.V., Jha D., Wozniak G.G., Bridgers J.B., Kerschner J.L. et al. . Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev. 2015; 29:1795–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y., Sabari B.R., Panchenko T., Wen H., Zhao D., Guan H., Wan L., Huang H., Tang Z., Zhao Y. et al. . Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell. 2016; 62:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y., Wen H., Xi Y., Tanaka K., Wang H., Peng D., Ren Y., Jin Q., Dent S.Y., Li W. et al. . AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 2014; 159:558–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schulze J.M., Wang A.Y., Kobor M.S.. Reading chromatin: insights from yeast into YEATS domain structure and function. Epigenetics. 2010; 5:573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dhalluin C., Carlson J.E., Zeng L., He C., Aggarwal A.K., Zhou M.M.. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999; 399:491–496. [DOI] [PubMed] [Google Scholar]
- 14. Zeng L., Zhang Q., Li S., Plotnikov A.N., Walsh M.J., Zhou M.M.. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010; 466:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andrews F.H., Shanle E.K., Strahl B.D., Kutateladze T.G.. The essential role of acetyllysine binding by the YEATS domain in transcriptional regulation. Transcription. 2016; 7:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andrews F.H., Shinsky S.A., Shanle E.K., Bridgers J.B., Gest A., Tsun I.K., Krajewski K., Shi X.B., Strahl B.D., Kutateladze T.G.. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 2016; 12:396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Erb M.A., Scott T.G., Li B.E., Xie H., Paulk J., Seo H.S., Souza A., Roberts J.M., Dastjerdi S., Buckley D.L. et al. . Transcription control by the ENL YEATS domain in acute leukaemia. Nature. 2017; 543:270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wan L., Wen H., Li Y., Lyu J., Xi Y., Hoshii T., Joseph J.K., Wang X., Loh Y.E., Erb M.A. et al. . ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature. 2017; 543:265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao D., Guan H.P., Zhao S., Mi W.Y., Wen H., Li Y.Y., Zhao Y.M., Allis C.D., Shi X.B., Li H.T.. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016; 26:629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calo E., Wysocka J.. Modification of enhancer chromatin: what, how, and why?. Mol. Cell. 2013; 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Dieuleveult M., Yen K., Hmitou I., Depaux A., Boussouar F., Bou Dargham D., Jounier S., Humbertclaude H., Ribierre F., Baulard C. et al. . Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016; 530:113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doyon Y., Cote J.. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004; 14:147–154. [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossetto D., Cramet M., Wang A.Y., Steunou A.L., Lacoste N., Schulze J.M., Cote V., Monnet-Saksouk J., Piquet S., Nourani A. et al. . Eaf5/7/3 form a functionally independent NuA4 submodule linked to RNA polymerase II-coupled nucleosome recycling. EMBO J. 2014; 33:1397–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boudreault A.A., Cronier D., Selleck W., Lacoste N., Utley R.T., Allard S., Savard J., Lane W.S., Tan S., Cote J.. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003; 17:1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burgess R.J., Zhou H., Han J., Zhang Z.. A role for Gcn5 in replication-coupled nucleosome assembly. Mol. Cell. 2010; 37:469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu W.H., Alami S., Luk E., Wu C.H., Sen S., Mizuguchi G., Wei D., Wu C.. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 2005; 12:1064–1071. [DOI] [PubMed] [Google Scholar]
- 30. Wu W.H., Wu C.H., Ladurner A., Mizuguchi G., Wei D., Xiao H., Luk E., Ranjan A., Wu C.. N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J. Biol. Chem. 2009; 284:6200–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fuda N.J., Ardehali M.B., Lis J.T.. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009; 461:186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nourani A., Utley R.T., Allard S., Cote J.. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 2004; 23:2597–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Santisteban M.S., Kalashnikova T., Smith M.M.. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000; 103:411–422. [DOI] [PubMed] [Google Scholar]
- 34. Buschbeck M., Hake S.B.. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 2017; 18:299–314. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Q., Zeng L., Zhao C.C., Ju Y., Konuma T., Zhou M.M.. Structural insights into histone crotonyl-lysine recognition by the AF9 YEATS domain. Structure. 2016; 24:1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cai Y., Jin J., Florens L., Swanson S.K., Kusch T., Li B., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W.. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 2005; 280:13665–13670. [DOI] [PubMed] [Google Scholar]
- 37. Doyon Y., Selleck W., Lane W.S., Tan S., Cote J.. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004; 24:1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.