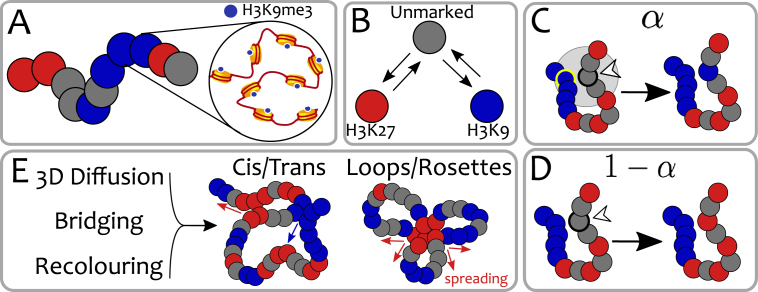
Figure 1.
Polymer model with dynamic epigenetic patterns. (A) In our coarse-grain polymer model, each bead represents a group of nucleosomes and its colour captures the predominant epigenetic mark. (B) Epigenetic marks are dynamic. They can change between red, blue or grey (no mark) according to biophysical rules. For example, a red bead can be thought of as an inactive Polycomb state (marked by H3K27me3) while a blue bead as a heterochromatic segment (marked by H3K9me3). The precise nature of the marks does not affect the qualitative behaviour of this generic model. In the Voter-like dynamics, each bead must go through the unmarked state (gray) before changing to the opposite colour (26). Each bead is selected at rate kR (see text and SM) and, (C) with probability α, it changes its colour ‘closer’ to that of a randomly chosen 3D-proximal bead (in this case the one circled in yellow, see also SM). (D) The same bead has probability 1 − α to undergo a random colour conversion (in this case to red, see SM). (E) The synergy between 3D chromatin dynamics, bridging due to (implicit) binding-proteins/TFs and epigenetic recolouring gives rise to dynamic structures such as loop/rosettes and cis/trans contacts which drive (cis and trans) epigenetic spreading (indicated by red/blue arrows, see text).