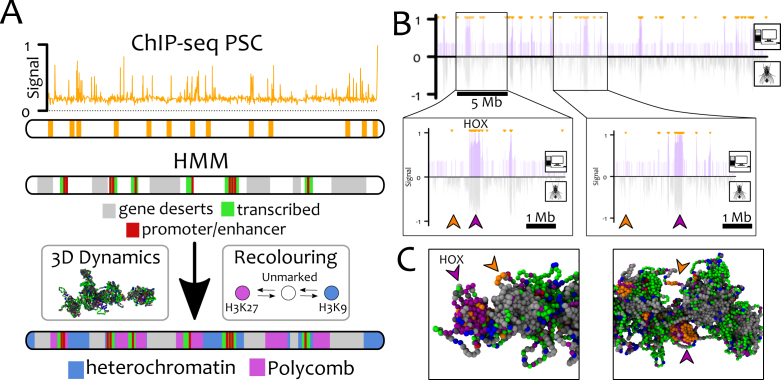
Figure 6.
GBM alone is able to recapitulate the distribution of polycomb marks in Drosophila S2 cells. Here, we perform chromosome-wide simulations of Ch3R of Drosophila S2 cells at 3 kb resolution (L = 9302) with GBM. (A) The location of PSC/PRE (bookmarks) are mapped onto beads using ChIP-Seq data from Ref. (54). Using the ‘9-states’ HMM data (60), gene deserts (regions lacking any mark in ChIP-seq data, state 9), promoter/enhancers (state 1) and transcriptionally active regions (states 2–4) are permanently coloured grey, red and green, respectively. The remaining beads (∼20%) are initially unmarked (white) and may become either heterochromatin (blue) or polycomb (purple). (B) In silico ChIP-seq data for H3K27me3 (top half, purple lines) is compared with in vivo ChIP-seq (60) (bottom half, grey line). Small orange arrows at the top of the profile indicate the location of the bookmarks. The excellent quantitative agreement between the datasets is captured by the Pearson correlation coefficient ρ = 0.46 – to be compared with ρ = 0.006 obtained between a random and the experimental datasets. We highlight that not all the bookmarked beads foster the nucleation of H3K27me3 domains (see big purple/orange arrowheads in the insets, corresponding to the HOX cluster). The reason can be found by analysing the 3D conformations of the chromosome (C). The non-nucleating bookmarks (orange arrowheads), although near in 1D, are found far from potential target beads in 3D space (purple arrowheads) and so fail to yield large H3K27me3 domains. See also Suppl. Movie 6 for a direct comparison of the results with and without bookmarks.