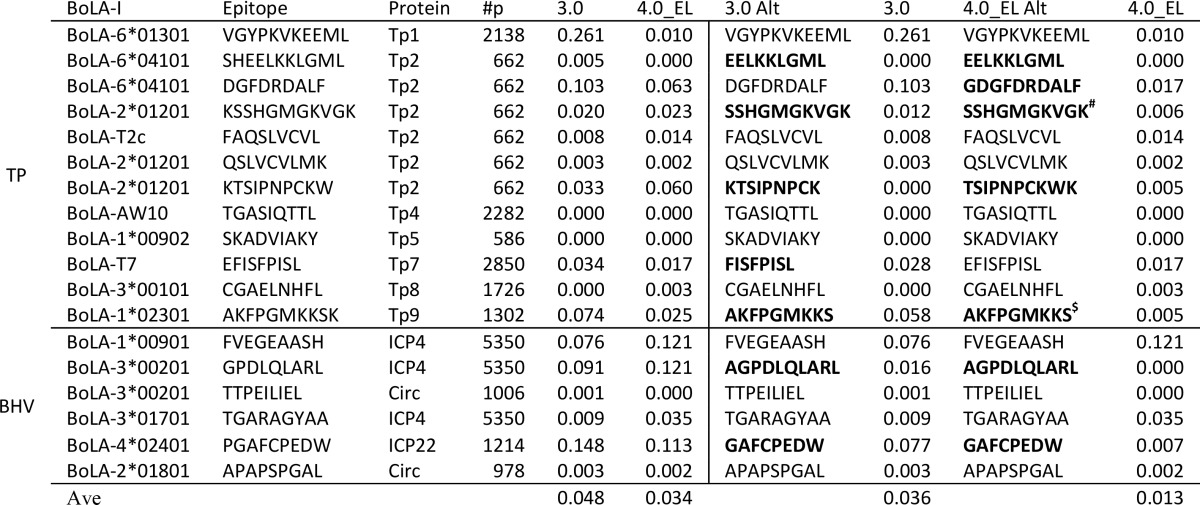
Table 3. Predictive Performance of the NetMHCpan-4.0 Eluted Ligand Likelihood Prediction Model (4.0_EL) Compared with NetMHCpan-3.0 (3.0) on a Data Set of Known BoLA-I Restricted T-Cell Epitopes from Theileria parva (TP) and Bovine Herpes Virus (BHV)a,b.
The part of the Table to the left of the vertical line gives the performance of the two methods on the original epitope data. The part of the Table to the right of the vertical line gives the results allowing each prediction method to suggest alternative epitopes overlapping with the known epitopes (either contained within known epitopes or with single amino acid extensions). In bold is highlighted the case where the two methods suggest alternative optimal epitopes.
#: Minimal epitope defined in ref (30); $: Minimal epitope (N. MacHugh personal communication).