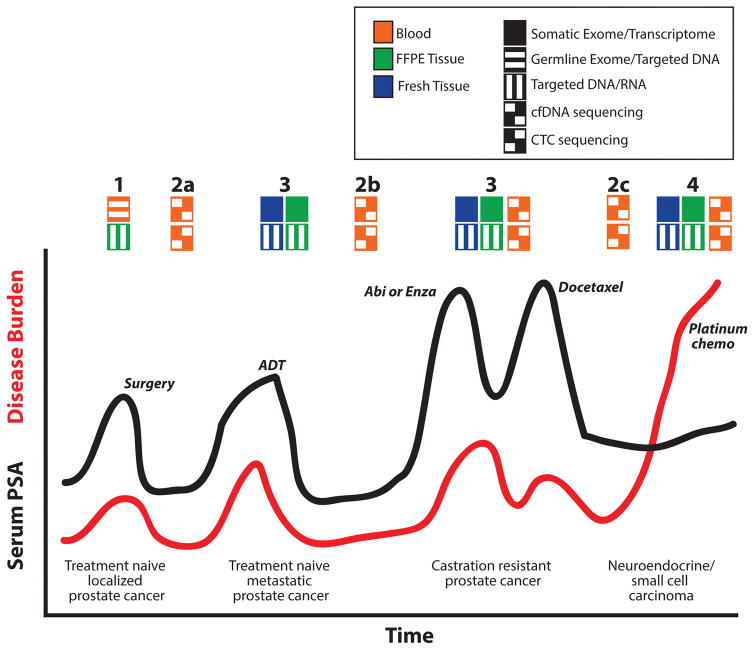
Figure 1.
Potential clinical utility of next generation sequencing (NGS) during prostate cancer progression. A timeline of serum PSA (black line) and disease burden (red line) along with treatments (italics) are shown for a hypothetical patient who progresses from localized untreated prostate cancer diagnosed and treated by radical prostatectomy to untreated treatment naïve metastatic prostate cancer to castration resistant prostate cancer (CRPC) and eventually neuroendocrine/small cell carcinoma. Opportunities for NGS to guide clinical management are shown above the graph according to the biocompartment assessed (color of the box) and NGS approach (pattern of the box) as indicated in the legend. At diagnosis (1), germline NGS assessment may be utilized to identify predisposing germline variants that may inform on later therapy and identify hereditary predisposition. Likewise, targeted DNA and RNA based assessment of FFPE biopsy and/or prostatectomy tissues may be used for prognosis and assessment of presumed clonal alterations that can be tracked and/or targeted during progression. NGS of cfDNA and/or CTCs isolated from blood may be used for non-invasive assessment of disease recurrence (2a) and assessment of clonal dynamics upon treatment. Diagnosis of metastatic disease by biopsy enables targeted DNA and RNA assessment of FFPE tissue (or comprehensive assessment if fresh tissue is obtained [most likely in the translational research setting]), and may have utility in predicting response to ADT or enrollment on clinical trials in the castration sensitive space (3). In addition to monitoring for development of CRPC after ADT (2a), NGS of cfDNA and/or CTCs may have particular utility for predicting response to second generation anti-androgens (such as abiraterone [abi] or enzalutamide [enza]) based on assessment of AR amplifications, mutations, or splice variant expression. Likewise, targeted or comprehensive NGS of CRPC biopsy tissue may have utility for identifying resistance mechanisms, novel targetable alterations, and identification of alterations enabling enrollment on umbrella and/or basket studies (3). NGS assessment of cfDNA and/or CTCs may be useful as a non-invasive complement to serum PSA to identify the development of AR independent clones (2c) and neuroendocrine/small cell prostate carcinoma when serum PSA may not be an accurate measurement of disease burden. Lastly, NGS of neuroendocrine/small cell prostate carcinoma (4) tissue may identify potential novel targetable alterations that developed during progression.