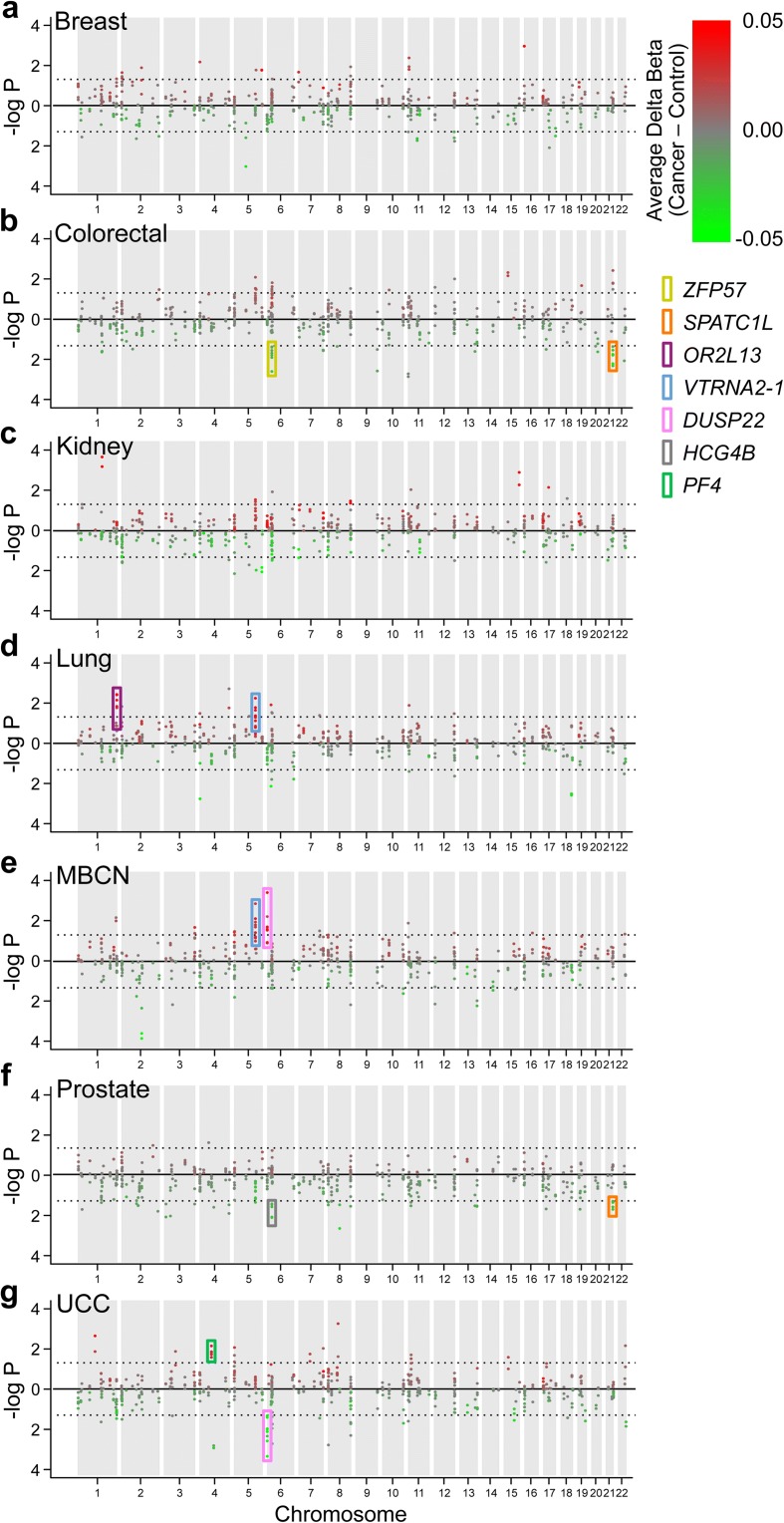
Fig. 7.
At clusters of probes showing epigenetic supersimilarity (ESS), peripheral blood methylation at baseline is associated with risk of later cancer. Manhattan plots illustrating results of conditional logistic regression analyses of the association between baseline probe-specific methylation (HM450) and risk of later a breast cancer, b colorectal cancer, c kidney cancer, d lung cancer, e mature B-cell neoplasm, f prostate cancer, and g urothelial cell carcinoma. Only probes within clusters of ≥ 2 probes are shown. Probes plotted with positive values (red) have positive coefficients (i.e., more methylation in cases than controls) and probes plotted with negative values (green) have negative coefficients (delta beta value scale indicated). The dotted lines indicate P = 0.05. Among the ten most CpG-rich ESS clusters, colored boxes indicate seven at which methylation is significantly associated with later cancer: ZFP57 (colorectal cancer, P = 0.008), SPATC1L (colorectal cancer, P = 0.009, and prostate cancer, P = 0.01), OR2L13 (lung cancer, P = 0.010), VTRNA2-1 (lung cancer, P = 0.025, and MBCN, P = 0.009), DUSP22 (MBCN, P = 0.001, and UCC, P = 0.001), HCG4B (prostate cancer, P = 0.007), and PF4 (UCC, P = 0.013)