Fig. 1. Principle of AlphaLISA™ screen for quantifying PRMT5 methylation activity.
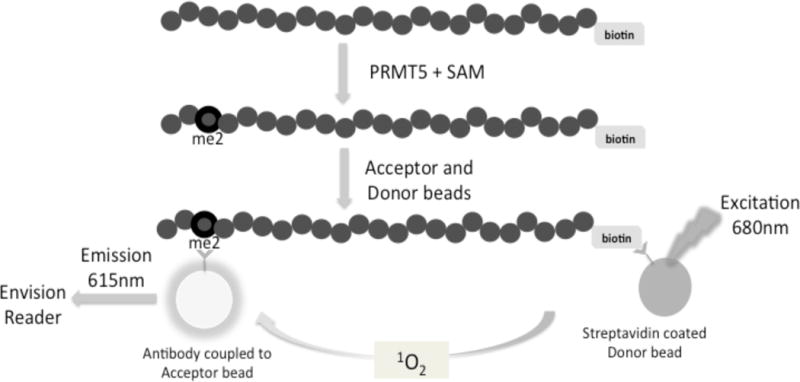
Biotinylated histone H4 is incubated with PRMT5 and a methyl donor, S-adenosyl-l-methionine (SAM). PRMT5 symmetrically dimethylates the third arginine (R3) on Biotin-H4 to form dimethylated Biotin-H4R3 (Biotin-H4R3me2) peptide. This product will further bind to Acceptor beads specific to the methylation site (Biotin-H4R3me2). Streptavidin-coated Donor beads will bind to the biotinylated substrate. Interaction between the Acceptor and Donor beads emits a chemiluminescent signal, which will be detected using an EnVision® reader. The methylation activity of PRMT5 is proportional to the intensity of the signal.
