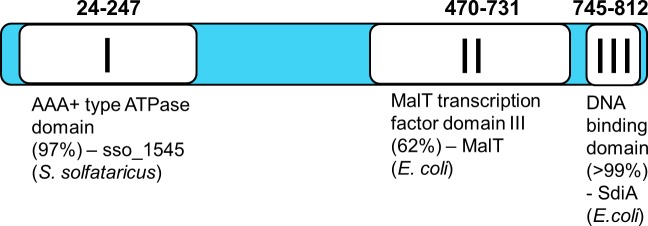
Fig 5. Overview of the three domains that can be found in iniR.
Noted domains are the most significant hits using Phyre2 structure prediction [21]. The number between brackets indicates the confidence with which the Phyre2 prediction was made. A combination of NCBI BLAST and the Phyre2 server was used to obtain domain predictions. Domain I (amino acid 24 to 247) is an AAA+ ATPase domain sharing 15% amino acid identity with sso_1545 in S. solfataricus. Domain II (470–731) is 16% identical to domain III of MalT in E. coli and the C-terminal domain (745–812) contains a DNA binding domain sharing 20% identity to E. coli SdiA. Displayed domains are proportional to actual domain size.