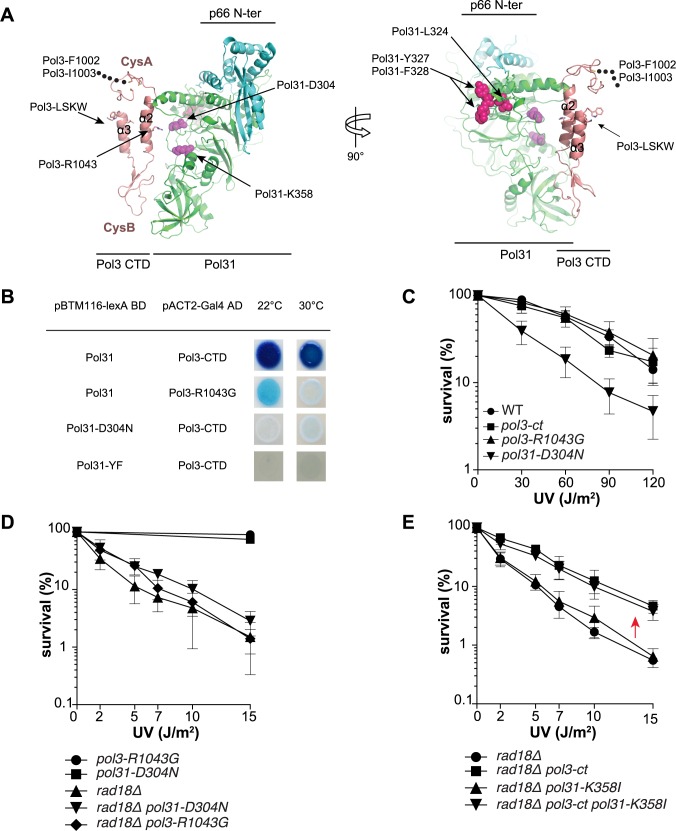
Fig 5. Functional analysis of the interaction between Pol3-CTD and Pol31 in cell survival after UV irradiation.
(A) Structural model of the Pol3-CTD/Pol31/P66 complex. Pol3 CTD contains two conserved cysteine-rich metal-binding motifs (CysA and CysB) that are separated by two α-helices. The Arg1043 residue of Pol3 lies within helix α2 between the Pol3 LSKW residues and Pol31. In addition, Pol31 Asp304 is very close to Pol31 interacting surface opposite to Pol3 Arg1043. The position of the LSKW motif at the C-terminal end of Pol 3 CTD is indicated by Pol3-LSKW. The conserved L1094 and W1097 residues of this motif are indicated by sticks. This motif is deleted in the pol3-ct mutant. The Pol3 residue R1043 within helix α2 is also indicated by sticks. The Pol3 residues F1002 and I1003 are located five amino-acids upstream of C1009 (the first amino-acid included in the structural model). These five amino-acids are indicated by black dots to show the proximity of Pol3-F1002 and -I1003 with Pol3-C1009. Pol31-D304 and -K358 residues are highlighted as magenta spheres. Pol31-L324, -Y327 and -F328 residues are shown as red spheres. Note that Pol31-L324 is buried in our model and not available for intermolecular interactions. (B) Yeast two-hybrid assays were performed using pBTM116 plasmids carrying WT and mutated Pol31 fused to the lexA binding domain (BD) and pACT2 plasmids carrying WT or mutated Pol3 C-terminal amino acids 1032–1097 fused to the Gal4 activating domain (AD). Each spots illustrates an individual co-transformant obtained following transformation of CTY10-5d. β-galactosidase activity was tested using an overlay plate assay at 22°C and 30°C. (C) Survival curves obtained after UV-irradiation of Pol δ mutant strains in which the interaction between Pol3 CTD and Pol31 is impaired. (D and E) Survival curves obtained after UV-irradiation of PCNA ubiquitination-deficient Pol δ mutant strains. (D) The pol3-R1043G and pol31-D304N alleles do not suppress rad18Δ-associated UV sensitivity. (E) The pol31K358I allele does not affect the suppression of rad18Δ-associated UV sensitivity by pol3-ct (red arrow).