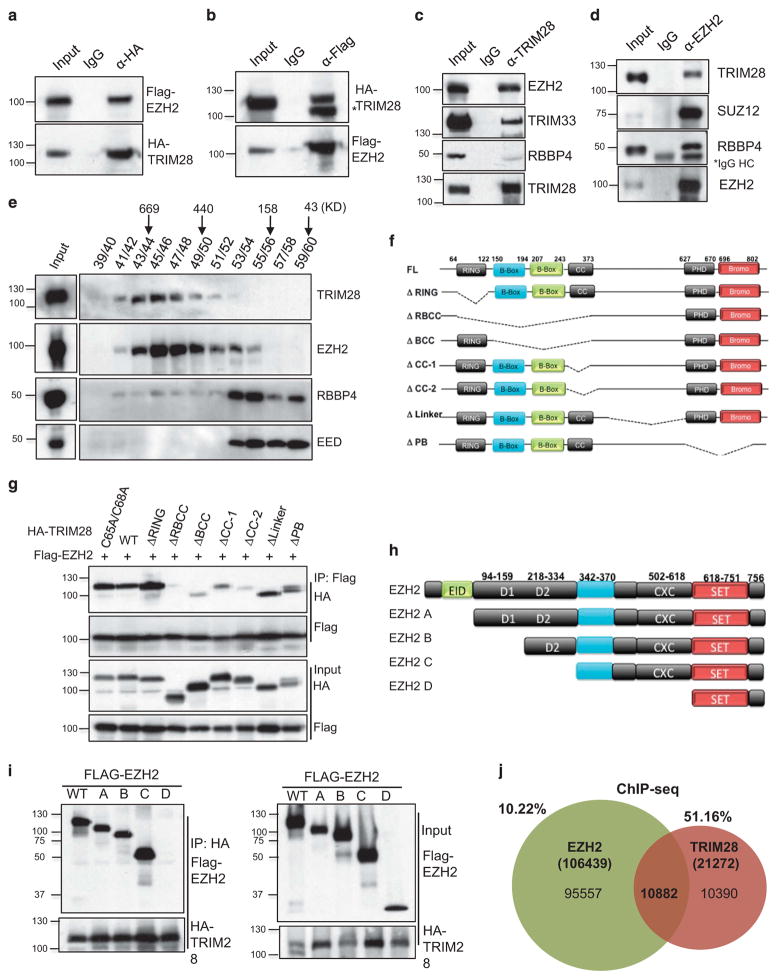
Figure 1.
The RBCC domain of TRIM28 interacts with the pre-SET region of EZH2. (a) Flag-EZH2 and HA-TRIM28 were co-transfected into HEK293T cells and either HA (a) or Flag was immunoprecipitated (b) followed by immunoblotting with Flag or HA antibody (*non-specific band). (c) Endogenous TRIM28 in MCF7 cells was immunoprecipitated and immunoblotted to detect EZH2. (d) Endogenous EZH2 in MCF7 cells was immunoprecipitated, and immunoblotted to detect TRIM28 and other PRC2 members. (e) Gel filtration assay. MCF7 lysate was fractionated by Superose 6 chromatography, followed by immunoblotting for EZH2, EED, RBBP4 and TRIM28. (f) Schematic representation of various deletion mutants of HA-TRIM28 used for co-interaction analyses. (g) Flag-EZH2 and HA-TRIM28 full-length and deletion mutants were ectopically expressed; followed by Flag-immunoprecipitations and immunoblotting with Flag and HA antibodies. Inputs are shown in bottom panels. (h) Schematic representation of various deletion mutants of Flag-EZH2 used for co-interaction analyses (i) HA-TRIM28 was co-transfected with various Flag-EZH2 mutated plasmids followed by HA-immunoprecipitations and immunoblotting with Flag and HA antibodies. Inputs are shown in panels on the right. (j) Venn diagram of intersection of genomewide EZH2-binding sites in HMEC cells with TRIM28-binding sites in MCF7 cells obtained from ChIP-Seq analysis.