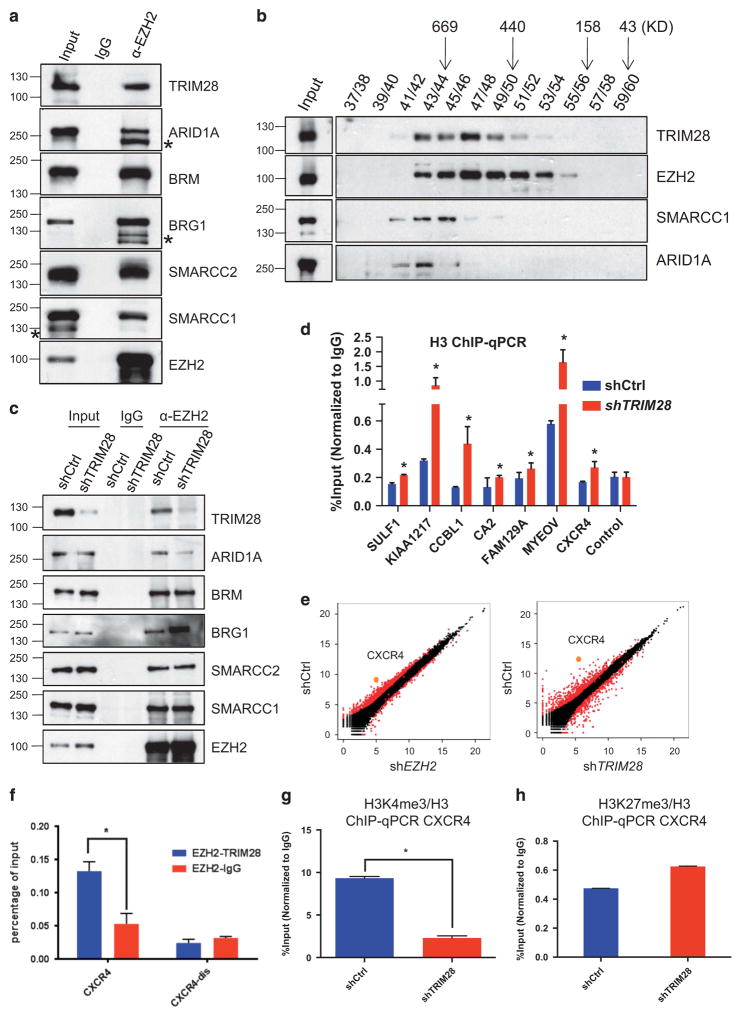
Figure 4.
TRIM28 and EZH2 regulate CXCR4 gene expression. (a) Endogenous EZH2 in MCF7 cells was immunoprecipitated, and immunoblotted to detect TRIM28 and members of SWI/SNF complex. (* non-specific band). (b) Gel filtration assay. MCF7 lysate was fractionated by Superose 6 chromatography, followed by immunoblotting for EZH2, SMARCC1, ARID1A and TRIM28. (c) Endogenous EZH2 was immunoprecipitated from lysates of shControl, shTRIM28 MCF7 cells, followed by western blotting using SWI/SNF complex members antibodies as indicated. (d) ChIP-qPCR analysis of histone H3 occupancy at selected gene promoters in shCtrl and shTRIM28 MCF7 cells. Error bars represent s.e.m. from three independent experiments (*P<0.05). (e) Scatter plots of gene expression levels measured as FPKM (fragments per kilobase of transcript per million mapped reads) values in shCtrl and shEZH2 (left), and in shCtrl and shTRIM28 (right) MCF7 cells. CXCR4 is highlighted as an example of TRIM28 and EZH2 co-activated gene (f) Sequential ChIP analysis in MCF7 cells: Chromatin fragments enriched by EZH2 were used in a sequential ChIP with antibody recognizing TRIM28 or IgG (negative control) at CXCR4 promoter and distal regions (CXCR4-dis), Error bars represent s.e.m. from three independent experiments (*P<0.05). (g, h) ChIP-qPCR analysis of H3K4me3 (g) and H3K27me3 (h) occupancy normalized to H3 at the promoter of CXCR4 gene.