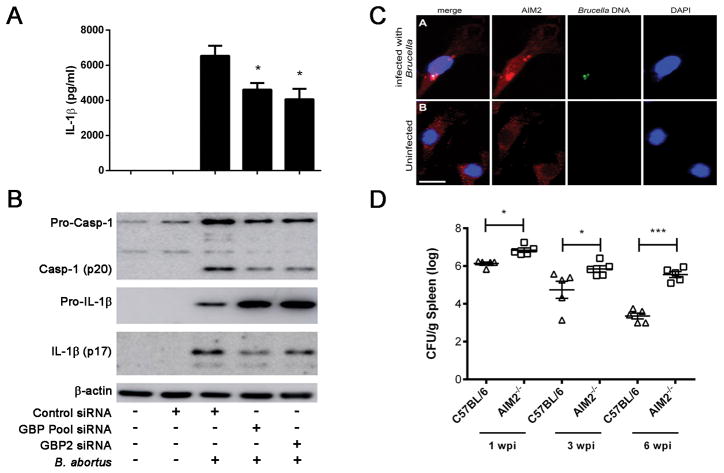
Figure 9. Inflammasome activation by Brucella partially requires functionally active guanylate binding proteins.
BMDMs from C57BL/6 mice were transfected with small interfering RNA (siRNA) from siGENOME smart pools (Dharmacon) for GBP2 and GBP pool (GBP2, 3 and 5) for 48 hrs and infected with B. abortus (MOI 100:1) for 17 hrs and IL-1β (A) secretion was measured by ELISA and pro-IL-1β (cell lysates), mature IL-1β (supernatant) and caspase-1 activation by Western blot (B). Significant differences from GBP2 and GBP pool siRNA in relation to siRNA control are denoted by an asterisk, two-way ANOVA (p<0.05). Data are representative of at least three independent experiments and three replicates in each experimental group. (C) Anti-AIM2 staining of Brucella-infected macrophages by confocal microscopy reveal aggregated speck formation of AIM2 in association with bacterial DNA. Brucella DNA was specifically labelled with click-IT kit and can be seen in green in the upper panel. DAPI staining allows visualization of bacterial DNA. Size bar shown corresponds to 20 μm in all panels. (D) C57BL/6 and AIM2 KO mice (n=5) were intraperitoneally inoculated with 106 CFU of B. abortus strain S2308. Residual B. abortus CFU in the spleen of wild-type and AIM2 KO mice were determined at 1, 3 and 6 weeks postinfection. Data are expressed as mean ± SD of five animals per time point and representative of three independent experiments. Significant difference in relation to C57BL/6 is denoted by an asterisk (p<0.05) or *** (p<0.001).